

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
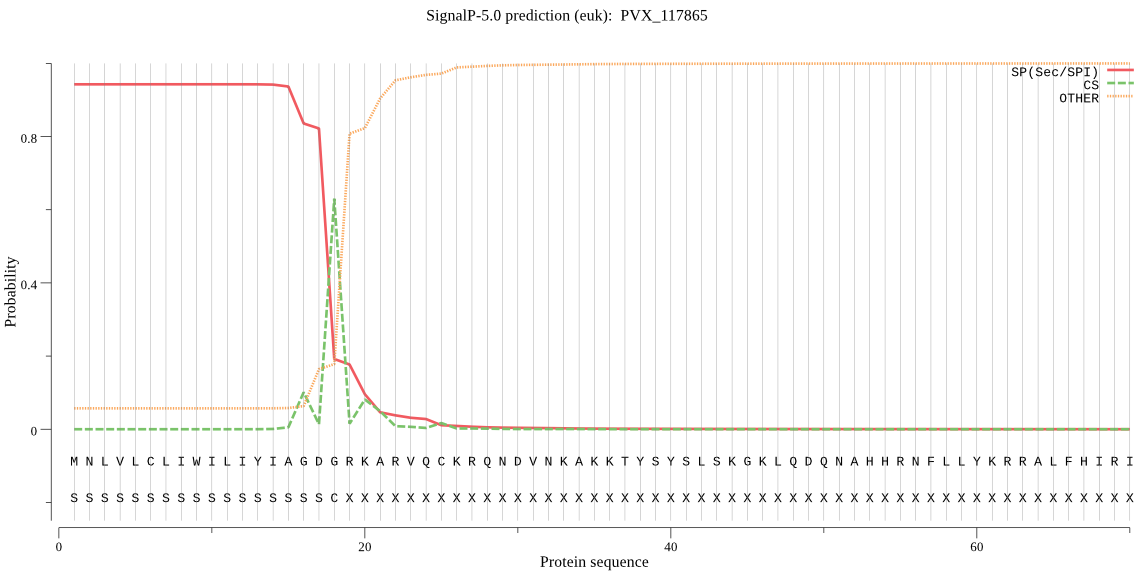
| PVX_117865 | SP | 0.042325 | 0.957088 | 0.000587 | CS pos: 18-19. GDG-RK. Pr: 0.5189 |
MNLVLCLIWILIYIAGDGRKARVQCKRQNDVNKAKKTYSYSLSKGKLQDQNAHHRNFLLY KRRALFHIRIPKKNLERQNLFSNDLRYKKINKFLENKKTNKLCYLHISNNNHVIKKKRNK NKTLQLLFEKKKLLQEYFSNLKSSFLDRYKNTKIVTKLFLSSSLLMLTLNLIGVKPEDIA LHDKRIIRAFEFYRIVTSALFYGDMSLYVLTNVYMLYVQSQELENSVGSSETLAFYLSQI SILSIICSYLKKPFYSTALLKSLLFVNCMLNPYQKSNLIFGINIYNIYLPYFSIFIDILH AQDLKASLSGILGVTSGSIYYLLNIYAYQKFNRRFFLIPRFLRNYLDSASVDDVL