

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
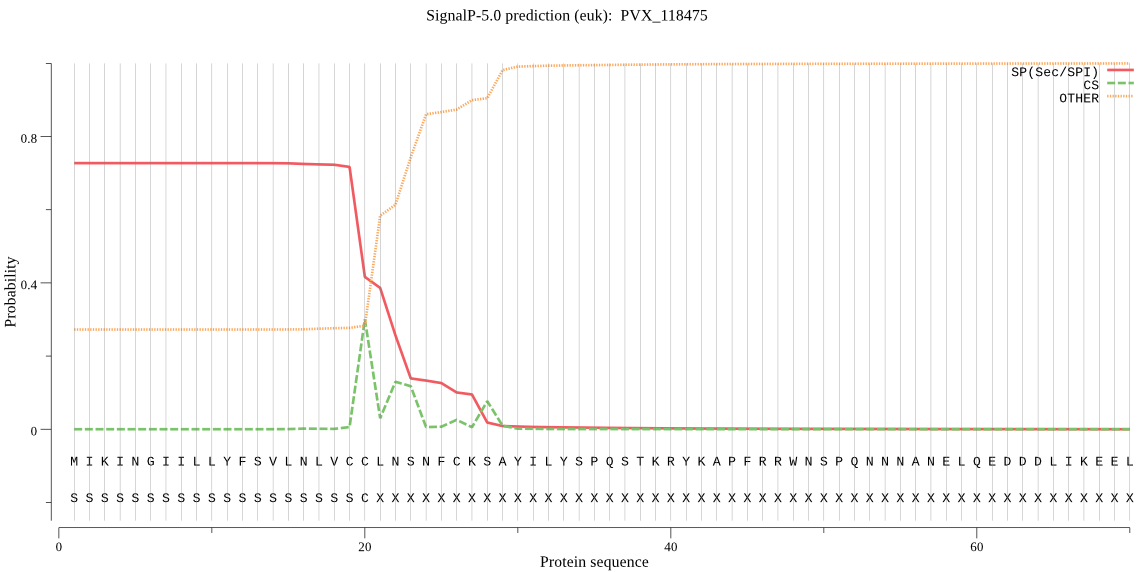
| PVX_118475 | SP | 0.110084 | 0.881513 | 0.008403 | CS pos: 20-21. VCC-LN. Pr: 0.5281 |
MIKINGIILLYFSVLNLVCCLNSNFCKSAYILYSPQSTKRYKAPFRRWNSPQNNNANELQ EDDDLIKEELPNRLTSIIYKKRKSKNKKKYILSNEKHVILPSHRISNEYKNYTNPIDYNY EEMHVYMEVNTGSVNEKKNQQGISHLCEHVSYMGSKNRKNIIDKNIRTNAYTDFHHIVFY ISVSLNKELYKENSLSHFQHDHIIKTVREDQIENYDIYTVDEFNYKHAILSKCIDTMVEV LKGETQFNRERIVKEKKAIFSEYSIINTVEYKMNSDIIKTLHKENRLSHRLPIGKLELLK RYEPKDIKKYFDLFFRTENVNLFIYGDVNVDIARKLVSEKFQNVKKEELTPEDMKYLAIL NDNTLRSRNKNLPAIVHMYGSDCSGKGHPESGPLNGSADEMGEVCGEEPIIGGADTGKEK IHTGRDGTPGDKKDTSEEVHKGSATPPPSGERSDDERSDDERPDDELPNDELPNDELPSD ELPSDERPDEERPDEERPDDNIADAHKLNELRSTLQNEALSELKFRAYLSEKYQPKLEEE KTLNKSGKKENPNLTTDFEIIKYSLNNVNINILMKEEIKSIRTMEDYKTYVMKDVMFYCL SFRFNVYKRDLFNGIDINEYTNINEGATIRTVEIKTTTKSFAEAVTAFYNFIKGLLAHGF SNDELQNYRANEIEYDDAGGGSGGGEGDCSSSSGAGTKQMKLVHPEKEEEEQILETRMNE MYTDEIQKIIDYNSCKHVYLNEKREKKLKQEIFDNLKIEQINSFAKNYFQYLFNIFKGNT NFKPNCVIIHVPKVDFTSFREDDVKRLFLDNINSVSDVPNFSLSFQNTLLSQRYLYEHIT TKLHMARYVQPGAGDPPRRDIFAGILRKIDQIKRDRQPFPCVLPGVETQTKINPIGTICT IDPIGTPGTPGTPTNMDTAPNLPLFRACYGMDGEDDLQTPQATHLRHPARGKKNTKLDQV PSELIMGATADRLNLKRYVAAEKSQREVENYQLANGIKVNIYKTQIDKKNIYLRLIIPHN DILKKKKKNVFPLLFSVICLFEGGEIESISRENVEIHCSNRSINICIDINDEYFFIDIYT YNKHENVNSAFSILNNILLQTRLEEAALKRVVDKLRKDFYEYKNNLQSFLLGQTLSYLSD GALGYQNFDLREAEEMSLETVQKVLRHLFSDPSLFELTIVGDTSDFVHYYVLHYIGTLPL RREASTNGKETIVNGVATTQRMNLHGDEKNPQQNDTPIGRDPLKEEYSLLCPLQNFEQKI KEATYVYLNEKEDHAVFLLIGKSANHFGFLSNGVHISLYLLQFLKKVLHHKMEDEHVEKN ITSEGFTQLVNHINVEKYKKEDITELITRKKKLYTNPIFFNAVSYIIQYILNSKLFHYLR EKKELTYDSSFEFINYEKYFAGFFTLLVQTNPKDLKIIKNEVLSSFENFTKNFYNYSDYL IENAKLSYLNKKTKDLKFFVDKISGMQLTHFPLKYKNKSLLRDNLILSRIEKIDVLLTLY VLFNQTAGYHISCGIASPKDVWAGVYRNINEL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_118475 | 436 S | KKDTSEEVH | 0.996 | unsp | PVX_118475 | 436 S | KKDTSEEVH | 0.996 | unsp | PVX_118475 | 436 S | KKDTSEEVH | 0.996 | unsp | PVX_118475 | 449 S | TPPPSGERS | 0.997 | unsp | PVX_118475 | 484 S | DELPSDERP | 0.995 | unsp | PVX_118475 | 798 S | VDFTSFRED | 0.994 | unsp | PVX_118475 | 37 S | YSPQSTKRY | 0.994 | unsp | PVX_118475 | 84 S | KKRKSKNKK | 0.994 | unsp |