

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_118620 | OTHER | 0.999970 | 0.000011 | 0.000019 |
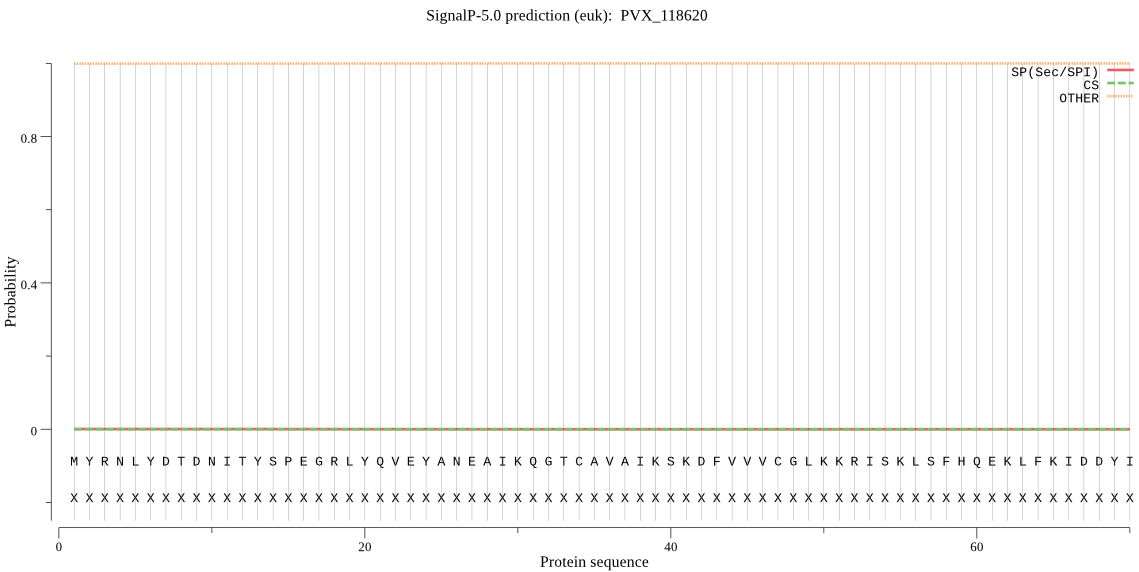
MYRNLYDTDNITYSPEGRLYQVEYANEAIKQGTCAVAIKSKDFVVVCGLKKRISKLSFHQ EKLFKIDDYIGVTMSGITSDAKVLTKYMRNECLSHKFLFDENMNIEKLVKKVADKYQQNT QRSSRRAFGVGLIIAGYFKEPYIFETKPNGSYFEYIALSFGARSHASKTYLEKNVHLFED SSLEELTIHCLKALRCSLSSENELTIENTSIAIVGKDKPWQEITTVDLVELLIRVNAEQR TENVDADIQNEGIPPNEVDASNPQGDQME