

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
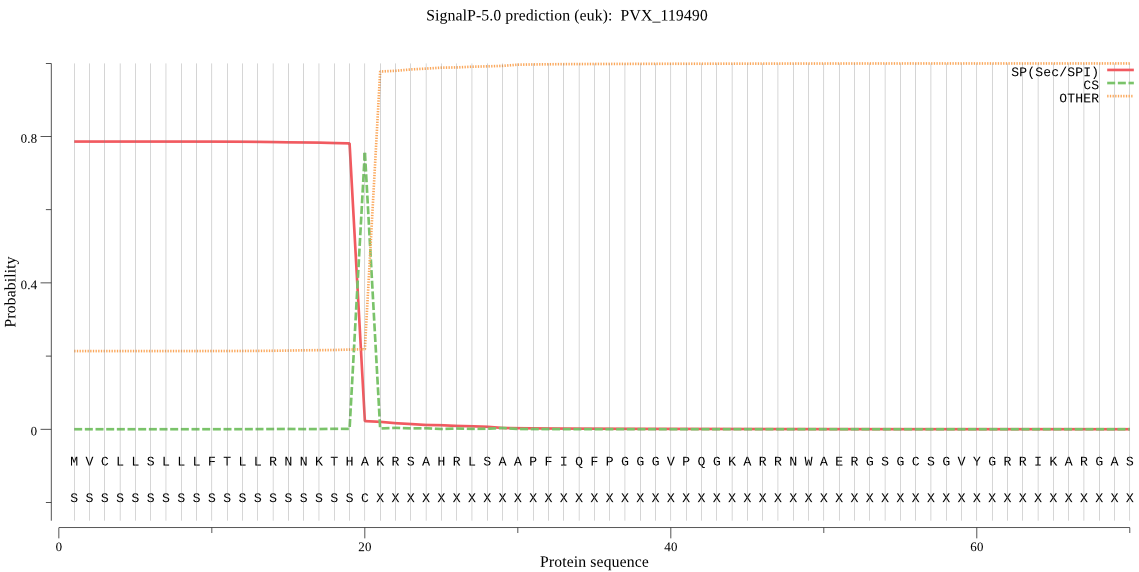
| PVX_119490 | SP | 0.308919 | 0.614260 | 0.076821 | CS pos: 20-21. THA-KR. Pr: 0.9633 |
MVCLLSLLLFTLLRNNKTHAKRSAHRLSAAPFIQFPGGGVPQGKARRNWAERGSGCSGVY GRRIKARGASSNSLPRSIGDDQECAAPGEESASHSAAERAAYGMAPSDPPAKGMKSVKNG ELRMDAATGVGGFLVGEGAHIEEPPPEGKQTHKGSSGRSGRSSSHRRYRRREGLLPRVNH IKDMKKDVKLFFFKKRIIYLTEEINKKTADELISQLLYLDSLNHDDIKIYINSPGGSINE GLAILDIFNYIQSDIQTISFGLVASMASVILASGKKGKRKSLPNCRIMIHQPLGNAFGHP QDIEIQTKEILYLKKLLYNYLSSFTNQTIETIEKDSDRDHYMNALEAKRYGIIDEVIQTK LPHPYFGGAVESGSG
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_119490 | T | 151 | 0.570 | 0.050 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_119490 | T | 151 | 0.570 | 0.050 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_119490 | 159 S | SSGRSGRSS | 0.993 | unsp | PVX_119490 | 159 S | SSGRSGRSS | 0.993 | unsp | PVX_119490 | 159 S | SSGRSGRSS | 0.993 | unsp | PVX_119490 | 163 S | SGRSSSHRR | 0.995 | unsp | PVX_119490 | 164 S | GRSSSHRRY | 0.997 | unsp | PVX_119490 | 336 S | IEKDSDRDH | 0.998 | unsp | PVX_119490 | 116 S | KGMKSVKNG | 0.996 | unsp | PVX_119490 | 156 S | HKGSSGRSG | 0.996 | unsp |