

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
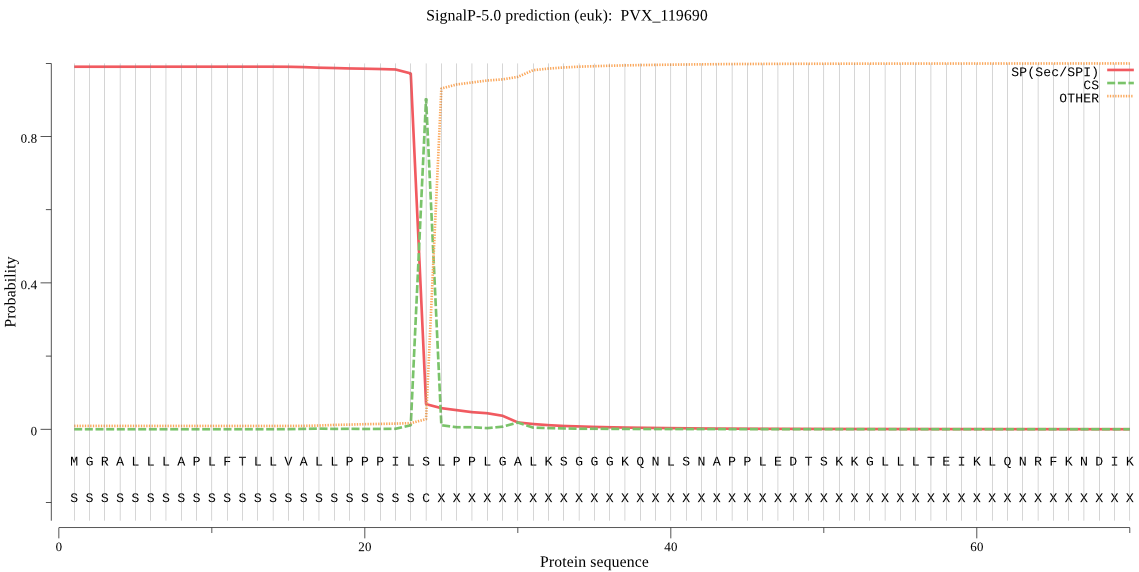
| PVX_119690 | SP | 0.001525 | 0.998360 | 0.000115 | CS pos: 24-25. ILS-LP. Pr: 0.8746 |
MGRALLLAPLFTLLVALLPPPILSLPPLGALKSGGGKQNLSNAPPLEDTSKKGLLLTEIK LQNRFKNDIKGFIRNVRSFHDSIEDRTPNSLLYVQEDLLNFHNSQFIGDIQIGTPPQSFR VVFDTGSSNFALPSTKCVKGGCASHKKFNPDESRTYARQLKGNKESIYTYIQYGTGRSIL EHGYDDVNLKGLRIKHQSVGLAIEESLHPFSDLPFDGIVGLGFSDPDFSFKKGYGTPLIE TIKKQNLLKRNIFSFYVPKKLREPGSITFGRANSKYALEGRQVEWFPVISIYFWEINLID VQLSGKNLNMCENRKCRAAIDTGSSLLTGPSSLMQPLIEKLDLRKDCSNKSSLPNISFIL KNVEGKEVKFAFTPDDYILEETDEEDNSVQCVIGIMSLDVPAPRGPIFVFGNVFIRKYYS IFDNDHKLVGLIEARHDW
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_119690 | 82 S | SFHDSIEDR | 0.996 | unsp | PVX_119690 | 420 S | RKYYSIFDN | 0.995 | unsp |