

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_119812 | mTP | 0.090000 | 0.432158 | 0.477842 | CS pos: 30-31. NRF-AA. Pr: 0.4034 |
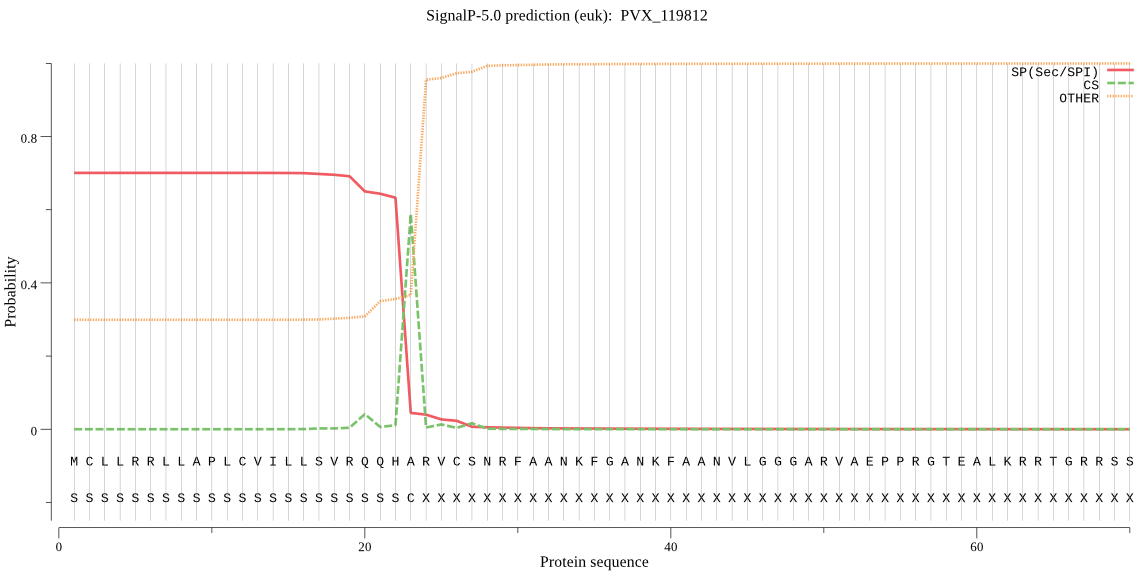
MCLLRRLLAPLCVILLSVRQQHARVCSNRFAANKFGANKFAANVLGGGARVAEPPRGTEA LKRRTGRRSSTPLNSDKKQKETPQHGYYFTKEKNKIVKKKKKKFSLNYCRDPKTTARGVK RRSHYPLLFISPPSTSLSEITESVKALFNVDYIENNYLYSYIKSFRRTPPITKLYLLCTL LLSICMHVNKNIYKLILFDFGKVFYEGQLWRLITPYFYIGNLYLQYFLMFNYLHIYMSSV EIAHYKNPEDLLTFLTFGYLSNLLFTIIGSMYSENVMNLQRYVNQLRRVILLGGKSSSTK GDTTVRIAKEQYNHLGYVFSTYILYYWSRINEGTLINCFELFLIKAEYVPFFFIIQNVLL YNEFSLFEVASILSSYFFFTYEKSLQLNFLRRFNLALLKALRVYPMYEAYREEYE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_119812 | 298 S | GKSSSTKGD | 0.992 | unsp | PVX_119812 | 298 S | GKSSSTKGD | 0.992 | unsp | PVX_119812 | 298 S | GKSSSTKGD | 0.992 | unsp | PVX_119812 | 70 S | GRRSSTPLN | 0.994 | unsp | PVX_119812 | 123 S | VKRRSHYPL | 0.994 | unsp |