

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_122425 | SP | 0.047322 | 0.952528 | 0.000150 | CS pos: 21-22. ALA-NY. Pr: 0.6525 |
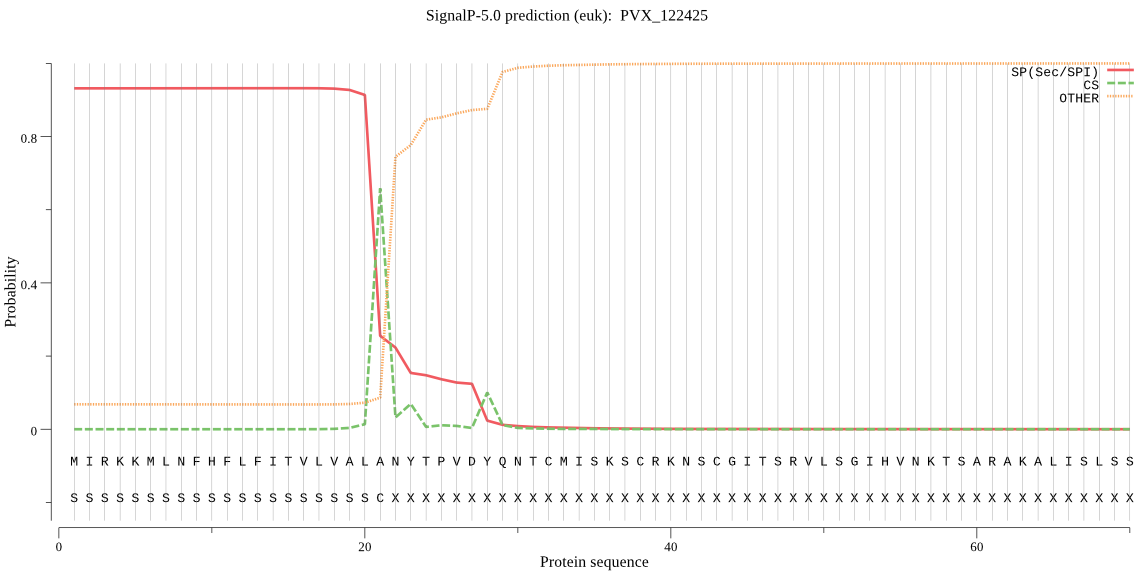
MIRKKMLNFHFLFITVLVALANYTPVDYQNTCMISKSCRKNSCGITSRVLSGIHVNKTSA RAKALISLSSLIYHLQLPKLVSLDLFRRDLFTGVKQKGKRSPVPSYIIQNRLMSENGDSG STNMSVTANQEKKRPGTGDASEGNNQSGISAAAQDKRMKGGDQSEEVSNVSGSTNAAMTN GASSTTEGGDNNNNGSGNDGKNEPKIHYRKDYKPSGFVIDNVTLNINIFDNETSVRSTLD MKLSEHYGGEDLIFDGVSLEIKEISIDNNKLMEGEHYKYDNEFLTIYSKFIPKGKFTFGS EVIIHPETNYALTGLYKSKNIIVSQCEATGFRRITFFIDRPDMMAKYDVTITADKEKYPV LLSNGDKLNEFEIPGGRHGARFNDPYLKPCYLFAVVAGDLKHLSDNYVTKFSKKNVELYV FSEEKYVSKLKWALECLKKAMKFDEDYFGLEYDLSRLNLVAVSDFNVGAMENKGLNIFNA NSLLASKKKSIDFSFERILTVVGHEYFHNYTGNRVTLRDWFQLTLKEGLTVHRENLFSEQ TTKTATFRLDHVDILRSVQFLEDSSPLAHPIRPESYVSMENFYTTTVYDKGSEVMRMYQT ILGDEYYKKGMDIYIKKNDGGTATCEDFNDAMNEAYKMKKGDKTANLDQYLLWFSQSGTP HVTAEYSYDAGKKEFVIEVTQVTNPDPNQKEKKALFIPIRVGFINPKNGQDVIPEVTLEF KKDKEKFIFNNVNEKPIPSLFRGFSAPVYIKDNLTDSERILLLKYDTDAFVRYNVCVDLY MKQILKNYQELLQAKSENKQESAEKPSLTPVSEDFINAIKYLMEDPHADAGFKSYIITLP RDRFILNYIKNVDTDVLADTKDFIYKQLGDKLNDLYFQMFKSLQAKADDMTHFEDESYVD FEQLNMRKLRNTLLTLLSRAKYPNMLDQIMEHSKSPYPSNWLASLAVSAYYDKYFDLYEK TYNQSKDDELLLQEWLKTVSRSDRKDIYDIIKKLETEVLKDSKNPNEIRAVYLPFTYNLR YFNDISGKGYKMMADIIMKVDKFNPMVATQLCDPFKLWNKLDQKRQDMMLNEMNRMLSME NISNNLKEYLLRLTNKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_122425 | 426 Y | SEEKYVSKL | 0.993 | unsp | PVX_122425 | 426 Y | SEEKYVSKL | 0.993 | unsp | PVX_122425 | 426 Y | SEEKYVSKL | 0.993 | unsp | PVX_122425 | 802 S | NKQESAEKP | 0.992 | unsp | PVX_122425 | 898 Y | EDESYVDFE | 0.992 | unsp | PVX_122425 | 1078 S | NRMLSMENI | 0.995 | unsp | PVX_122425 | 184 S | NGASSTTEG | 0.997 | unsp | PVX_122425 | 237 S | TSVRSTLDM | 0.99 | unsp |