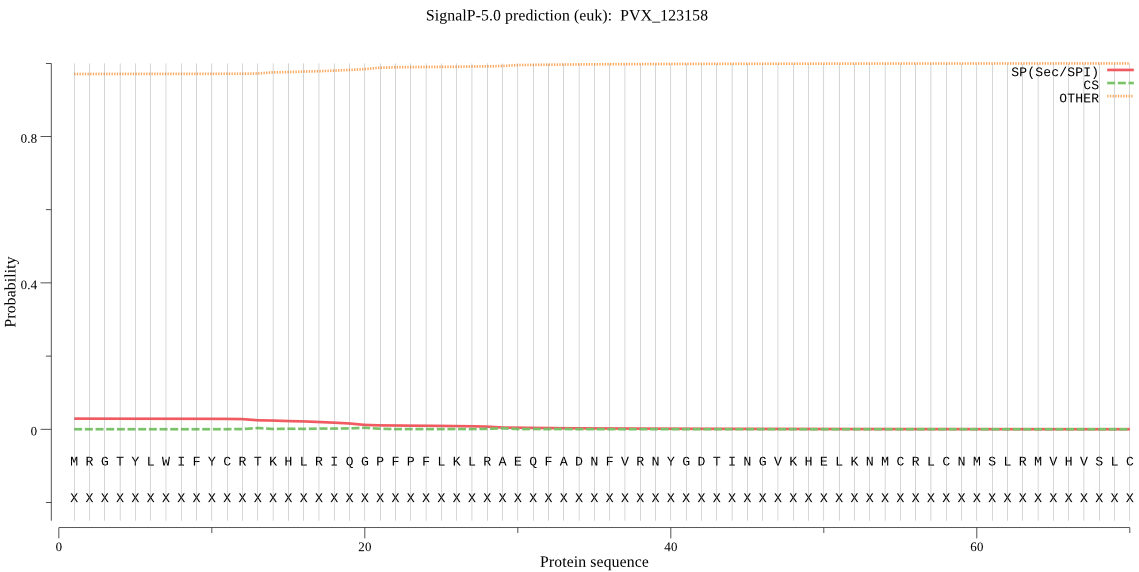
Fasta :-
MRGTYLWIFYCRTKHLRIQGPFPFLKLRAEQFADNFVRNYGDTINGVKHELKNMCRLCNM
SLRMVHVSLCESANGGGDGTADGCERTFESTFESTSDHTVGRTSEHSPEHRLSDGGHGAP
AESDQLEQNTQPEKNSRSKLATHLIIHYVKEYVKYIVHSLLHFCLHLCDAFYRSLSVLPN
GYLEFPKQFDYSSSISDNLLSSFVTIYKIYRQSNSSEFRFKLDQLAFLGSGFIYDQRGYV
LTAAHNITNTEGTFVIKNEDNFYVATVAGLHRESDVCVMKINSEEQFSHIPLDPSREFLK
PGEPVITYGQIQNFDKETCSVGVVNQPRQTFTKFESFCEKEQTCLYPFIQISNPINKGMS
GSPLIDKHGNLVGMIQKKIDNYGLALPVNILKNVTTHLQEEGTYKEPFLGIVLREKEQST
SADYKNCKNELKIQDVLANSPADVAGIAKGDIMLTINNKRIKNICDVHEILNSTSDGYIT
VDIIRHGKSKKIKVKM