

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_123162 | SP | 0.080226 | 0.911333 | 0.008442 | CS pos: 25-26. LCC-FV. Pr: 0.5375 |
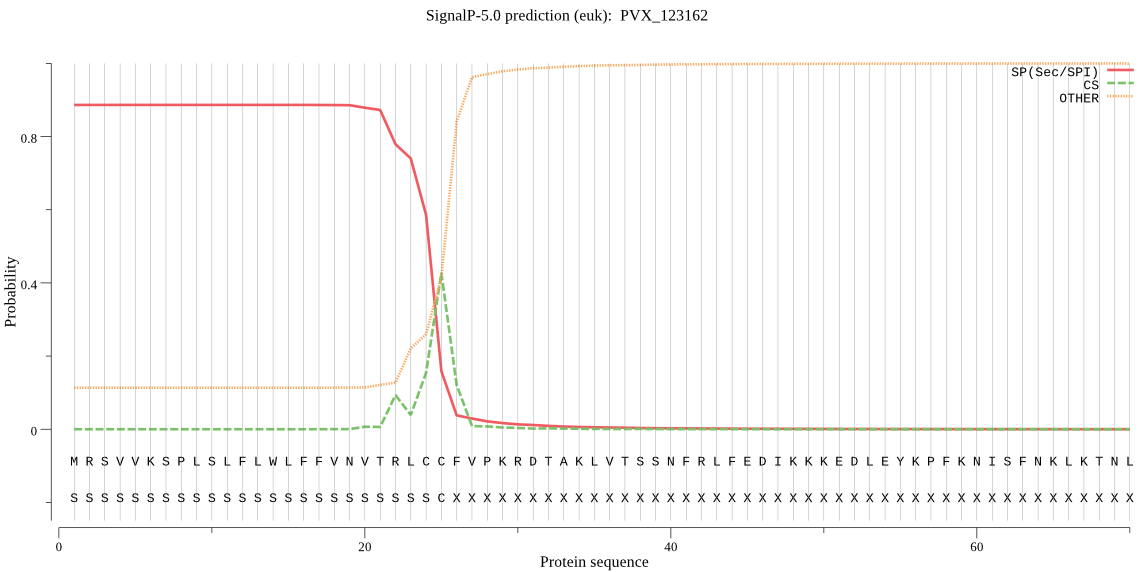
MRSVVKSPLSLFLWLFFVNVTRLCCFVPKRDTAKLVTSSNFRLFEDIKKKEDLEYKPFKN ISFNKLKTNLKNFLVINKKFNISDVKQVSFLGKFHNYEKIENYGVNEICILGRSNVGKST FLRNFIKYLINVNEHATVKVSKNSGCTRSINLYAFENGKKKKLFILTDMPGFGYAAGIGK KKMEFLMKNLEDYIFLRNQICLFFVLIDMSVDIQKVDVSIVDAIRKTNVPFRVICTKSDK FSASAEERLQAIKNFYQLEKIPIHISKFSTHNYINIFKEIQHHCNLDTP
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PVX_123162 | 244 S | KFSASAEER | 0.992 | unsp |