

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
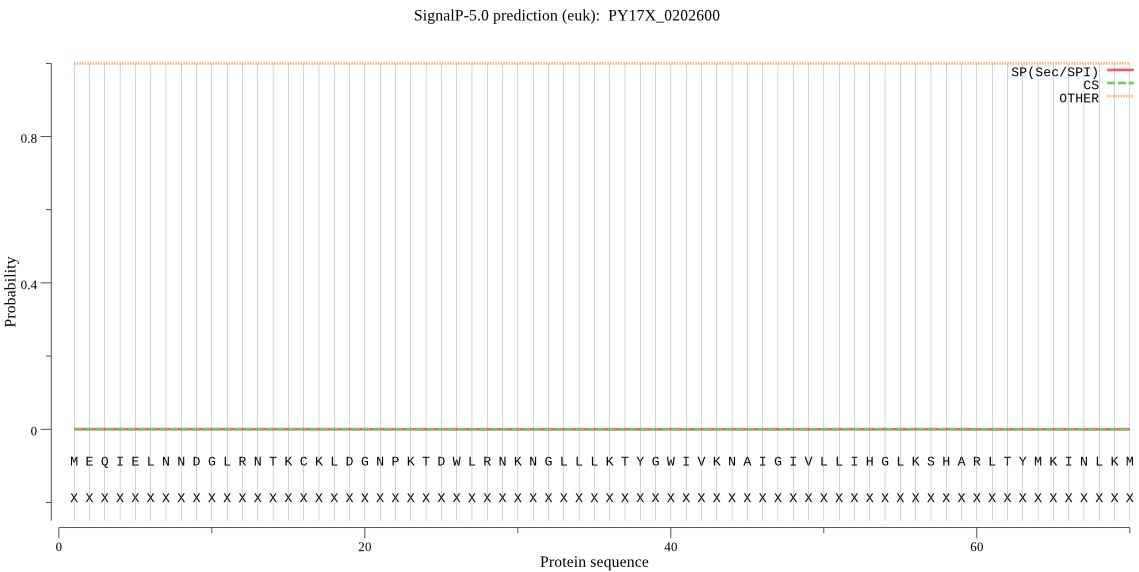
| PY17X_0202600 | OTHER | 0.999881 | 0.000057 | 0.000061 |
MEQIELNNDGLRNTKCKLDGNPKTDWLRNKNGLLLKTYGWIVKNAIGIVLLIHGLKSHAR LTYMKINLKMTNKNKSLVVDSNNYYIYKDSWIENFNKKGYSVYALDLQGHGESQAWGNIK GNFSSFDDLVDDVIQYMNKIHDEISNDNQTDDESTHIVTTKKKRLPMYIIGHSLGGGIAL RILQLLKKEQKDKINSEDANDNNMIEDIINDMDNSNDHAIGNMNNTHLITNSNDYDSDKS CASTSSTTNAIDGPSDKDEGCYNYLDKFNIKGCVTLSGMMRFKTIFDFGNNLIERFYLPA VDIMSRVAPHVLISSEIGYKRSKYVDNICKHDKFRNNSGIKFKCMYEVIKASITLNYNIN YMPTNVPLLFVHSKDDSICHYQGVISFHNQANVSKKKLHIVNGMNHAITVEQGNEQILKK ILEWISNLKRDDEIEDEINEINAEIEGEIEGEIEDEIEDETKNEI
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_0202600 | 255 S | IDGPSDKDE | 0.997 | unsp |