

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
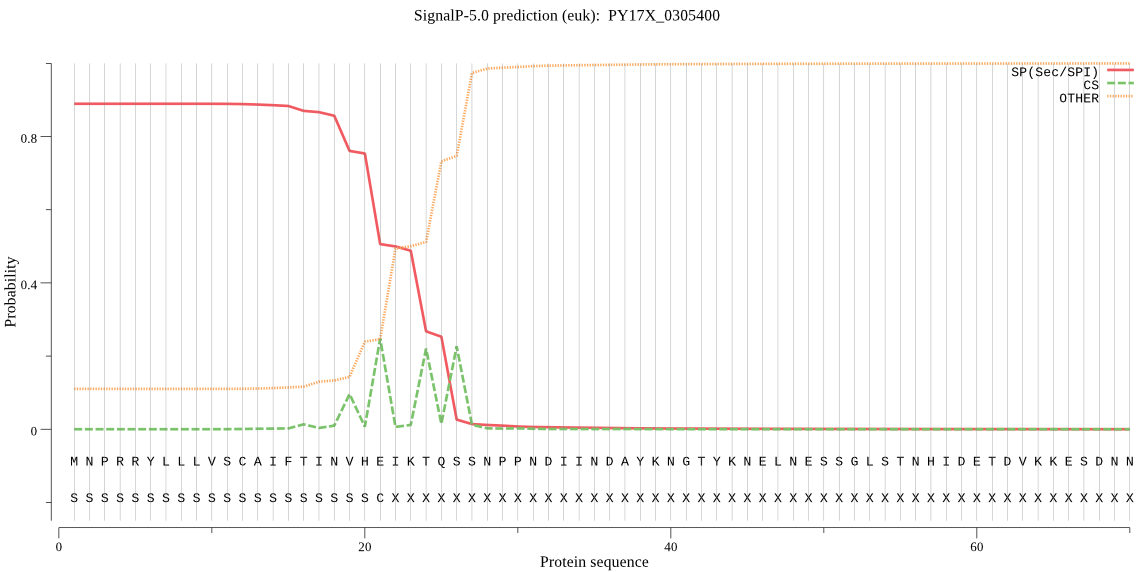
| PY17X_0305400 | SP | 0.040052 | 0.959537 | 0.000411 | CS pos: 24-25. IKT-QS. Pr: 0.5500 |
MNPRRYLLLVSCAIFTINVHEIKTQSSNPPNDIINDAYKNGTYKNELNESSGLSTNHIDE TDVKKESDNNTKNDSLNNEISDTETLIKSSLSKNGKGIKAIGVCNEDFRIYFGPYIWIYV TLSENIIKIEPENGVRTSINLNNLNNKCNDKKNDTFKFTAKIEEDILKIKWKTYTNETDQ EPKSDIKKFRLPDLAKDLTSIQIFTTNTKENIIESKTYDIDKNIPEKCSAISASCFLGGS LNIESCYHCTLLAQKYPNDDECFNYISSELKNKINNDTIIKGEDDLDENSDEKILREHIY KILKKMYNNDSDYCGNNRCNKKMIKNIKELDTDLQIYIKNYCDILKKVDKSGTLDSHEIA NEVEAFNNLVRLLNSHTSENIYILFEKLKSPAICVKNINEWIVRKRGLVISNEYDINSYD TTEEPNIKNILSDAFNEEDTEAIGNDIQNNEIDDENEIINLKSSSNKKLTSAYFNSSRYC NKDYCDRWQDKTSCISNIEVEEQGECGICWIFASKLHLETIRCMRGYGHYRSSALYVANC SDRKKSEICDIGSNPIEFIQILHDVKYLPLESDYPYSYYNVGDSCPTPKSNWTNLWGNNK LLYFKSGSVEFMSSYGFIAITSSNHLDDFDTYVQIIKNEIRNKGSVIAYIKTNNVIDYDF NGKIIHNLCGDDNDDADHAATIIGYGNYISNTGEKRSYWLIRNSWGYYWGDEGNFKVDIY GPQYCKYNFIQTVVFFKLDLGVIEGPQNNNQVYNYFSRHIPAFFTNLFYSSYDKRWDELS SDEELNNYNKNISISGQTDNQINTNNDFYNEDTDYINEISTSSSKNISIQKKLDITHVLK YVKNNKTQTSFIKYDDILEIEHEHNCSRVQSIGLKNKNECKSFCLENWVKCKNHPSPGYC LTTLYSGEDCFFCNI
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_0305400 | 546 S | DRKKSEICD | 0.995 | unsp |