

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_0305500 | SP | 0.000649 | 0.999351 | 0.000000 | CS pos: 21-22. AIS-NE. Pr: 0.9145 |
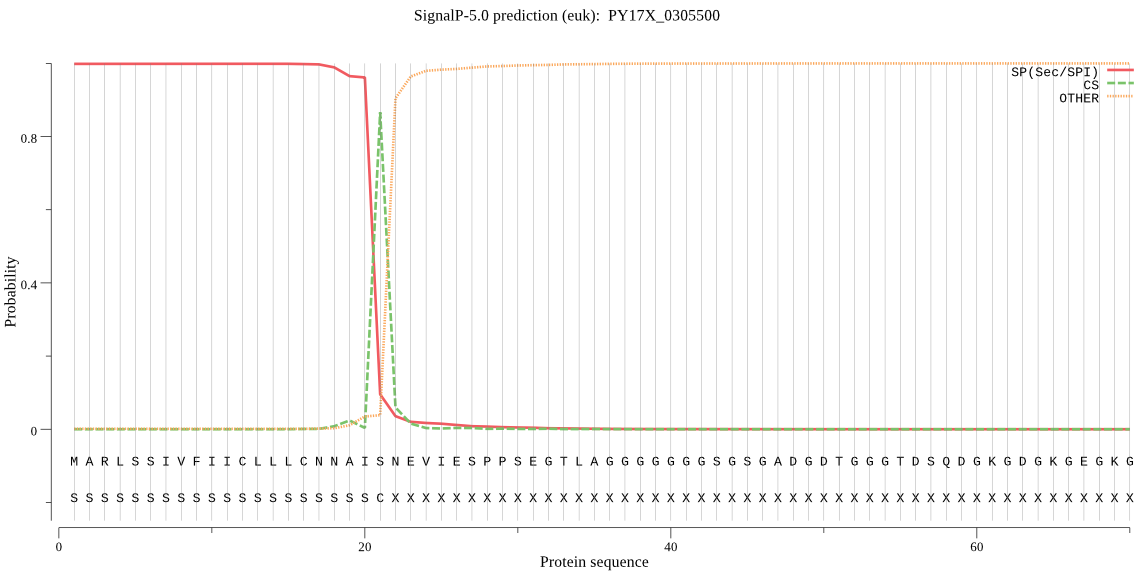
MARLSSIVFIICLLLCNNAISNEVIESPPSEGTLAGGGGGGGSGSGADGDTGGGTDSQDG KGDGKGEGKGEGKGETVGDKGKNNEGSQTDQELQTNPGSDQDSGSSGNKVPGAPSDDVNN PTGDLSTGLSGGKVTGASSDHVPGPTGEQGSDSKGGQKKEQPPPKEPAPTTPKEPAPTTP KEPEPTTPKEPEPTTPKEKAGDDPEKQVSPEGSASKTVEKKELSSSGDSLPSSSEDPGSP QKKPAGEEDPPSPPKKEESLPAKKPEAPPAKPAAPESLKVSQAGLDPNAQKQGQGKRPKR ALPPQVQVPQVNDITDMESYLLKNYDGVKVIGLCGVYFRVQFSPHLLLYGLTKFSIIQIE PFFERVRIDFEHQHPIKNKCAPGKAFAFISYIKDNILTLKWKVFTPPSDLFANEVVKEKI LSTVPEEPITEDESPVLVDVRKYRLPPLERPFTSIQVYKANTKQGLLETKNYILKNAIPE KCSKISMDCFLNGNVNIENCFKCTLLVQNAKPTDECFQYLPSDMKNNLDQIKISGQSDED TKENDLIESIEILLNSFYKVDKKTKKMSLITMDDFDDVLRTELFNYCKLLKELDTKKTLE NAELGNEIDIFNNMLRLLKTNEGETKLNLYKKLRNTAICLKDVNTWAEKKRGLILPEELT QDQMTEGQNEEPNDEDPDDRVDLLELFDDNQDENIVDKDGIIDMSMAIKYAKLKSPYFTS SKYCNYEYCDRWQDKTSCISNIDVEEQGNCSLCWLFASKLHIETIRCMRGYGHNRASALY VANCSERTGEEVCNDGSNPLEFLKILEKNKFLPLESNYPYLWKNVSGTCPNPQNDWTNLW GNTKLLYNNMYGQFIKHRGYIVYNSRFFAKNMDVFIDIVKREIRNKGSVIAYIKTQGVID YDFNGRYISNICGHSHPDHAVNIIGYGNYISDNGEKRSYWLIRNSWGYYWGHEGNFKVDV LGPDNCVHNVIHTAIVFKIDMEPDNASKNDDQLVDESDKSYFPQLSSNFYHSLYYNNFEG YEAKNDDENDQNYDKVDVSGQSEGNQTDLKNGQTDPQKTQTDRTVTQPGASPTENPGTAG SNLTTQNTGGSSPTVTDTSSPTTTVTETPGVTTSAATASIEKKIQILHVLKHIENYKMTR GLVKYDNLNDTKSDYACARSYAYDPKNHNECKQFCEENWERCKDHYSPGYCLTTLSGKNK CLFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0305500 | 179 T | PAPTTPKEP | 0.996 | unsp | PY17X_0305500 | 179 T | PAPTTPKEP | 0.996 | unsp | PY17X_0305500 | 179 T | PAPTTPKEP | 0.996 | unsp | PY17X_0305500 | 187 T | PEPTTPKEP | 0.995 | unsp | PY17X_0305500 | 195 T | PEPTTPKEK | 0.995 | unsp | PY17X_0305500 | 209 S | EKQVSPEGS | 0.997 | unsp | PY17X_0305500 | 225 S | KELSSSGDS | 0.996 | unsp | PY17X_0305500 | 232 S | DSLPSSSED | 0.992 | unsp | PY17X_0305500 | 233 S | SLPSSSEDP | 0.998 | unsp | PY17X_0305500 | 537 S | ISGQSDEDT | 0.998 | unsp | PY17X_0305500 | 1071 S | QPGASPTEN | 0.997 | unsp | PY17X_0305500 | 87 S | NNEGSQTDQ | 0.996 | unsp | PY17X_0305500 | 171 T | PAPTTPKEP | 0.996 | unsp |