

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
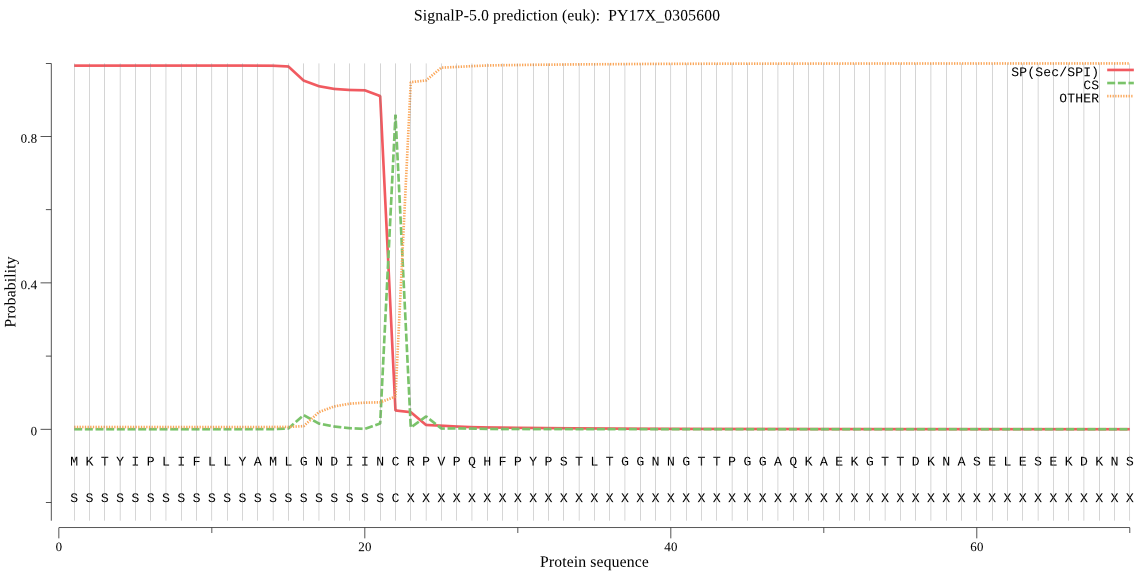
| PY17X_0305600 | SP | 0.048679 | 0.951320 | 0.000000 | CS pos: 22-23. INC-RP. Pr: 0.8956 |
MKTYIPLIFLLYAMLGNDIINCRPVPQHFPYPSTLTGGNNGTTPGGAQKAEKGTTDKNAS ELESEKDKNSQKSTDPSAPSSSNIATLPGSPDSAKQGSGSDSSDSSDSSDSSDSSDSSDS SDSSDSSDSSDSSDSSDVTRTKEVTPGTPVGTNTSDQQQSQDALQKAAGGPSSDPASISS SEVIVMDISKIKRRSEHRGTVESALLKNYKGVKVTGSCGDEVGIFLEPYIYISVDSKNNV MDLSATFPNVLGQHITIFSKESPMKNLCYDDDHLYYKMLVYLDQDVLTIKWTVFPIDPGM GKCELCANHNSENNVDVKKFRIPKLETPFTTIQVHSLLTKENQVVIKSKDYSLINDMPKK CDKIATKCFLDGKTDIEPCYTCSLLTENFPRTDECYNYSSPLVKEYNQVLAVGQSDEDNV DDKNLSLVDSINNILNDIYKTDEQNNKVLIDVEDLSTNLKKELTYYCQILNEIDTSGTLD IYKIGNSVEIFNNLIRLLKNHQDENKSYMINKLKNPAICMKHVEQWVVNRKGLKLPIILD DSETENKNSEESKNTNISIIYEEGPDGIIDLTKTIDGNTVSGLGVLANKLDAFCNNDFCD RLKDNNSCISNIDVEEQGNCATSWIFASKFHLETAICMKGHDNFTTSALYVANCSKKDPK DKCLTGSNPLEFLDILQANAYLPNESNYPYNYKNVGDSCPETNESWVNLFDKVKLENSND DTKSIIKGYTSYESKDYKSNMNEFVDIIKNKIKKLGSVIAYVKFSDFMGYDFNGNKVHKL SGSGTPDMIVNIIGYGKYINENNQVKPYWLVRNSWGKHWADEGNFKVDMETPENSEHNFI HTVLVFDLDLPVAQKINNDAPIYNYYLKNSPNFYKNLYYNHVSRKNAKAQADADAAISGQ AEEGPTKVEGVSNDKGQTLQSGGGSSQDGTANNVPGSQSGSGITGNGATVKDGATVQAAQ GQVANGQTGPGSAVTPSQTVQPSPGGSGLPPTGSDAGSRSPPTGTPSGSRFPPKGSPTVT VLTSSGGNGAQTQTNLNPASGSVQSNSATTAVNTESQANSTKNTEVFHVLKVVRNKKVQT SLIKYNKYNEMGEHSCSRAMSRDPLNKEKCIQFCSDNWDHCYFDLYPGYCLNKLKRDNEC NFCAV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0305600 | 70 S | KDKNSQKST | 0.997 | unsp | PY17X_0305600 | 70 S | KDKNSQKST | 0.997 | unsp | PY17X_0305600 | 70 S | KDKNSQKST | 0.997 | unsp | PY17X_0305600 | 93 S | GSPDSAKQG | 0.99 | unsp | PY17X_0305600 | 98 S | AKQGSGSDS | 0.996 | unsp | PY17X_0305600 | 105 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 108 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 111 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 114 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 117 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 120 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 123 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 126 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 129 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 132 S | DSSDSSDSS | 0.993 | unsp | PY17X_0305600 | 177 S | SDPASISSS | 0.991 | unsp | PY17X_0305600 | 195 S | IKRRSEHRG | 0.997 | unsp | PY17X_0305600 | 415 S | AVGQSDEDN | 0.991 | unsp | PY17X_0305600 | 426 S | DKNLSLVDS | 0.992 | unsp | PY17X_0305600 | 542 S | ILDDSETEN | 0.995 | unsp | PY17X_0305600 | 655 S | VANCSKKDP | 0.993 | unsp | PY17X_0305600 | 731 S | KGYTSYESK | 0.996 | unsp | PY17X_0305600 | 1060 S | SQANSTKNT | 0.996 | unsp | PY17X_0305600 | 60 S | DKNASELES | 0.995 | unsp | PY17X_0305600 | 64 S | SELESEKDK | 0.996 | unsp |