

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
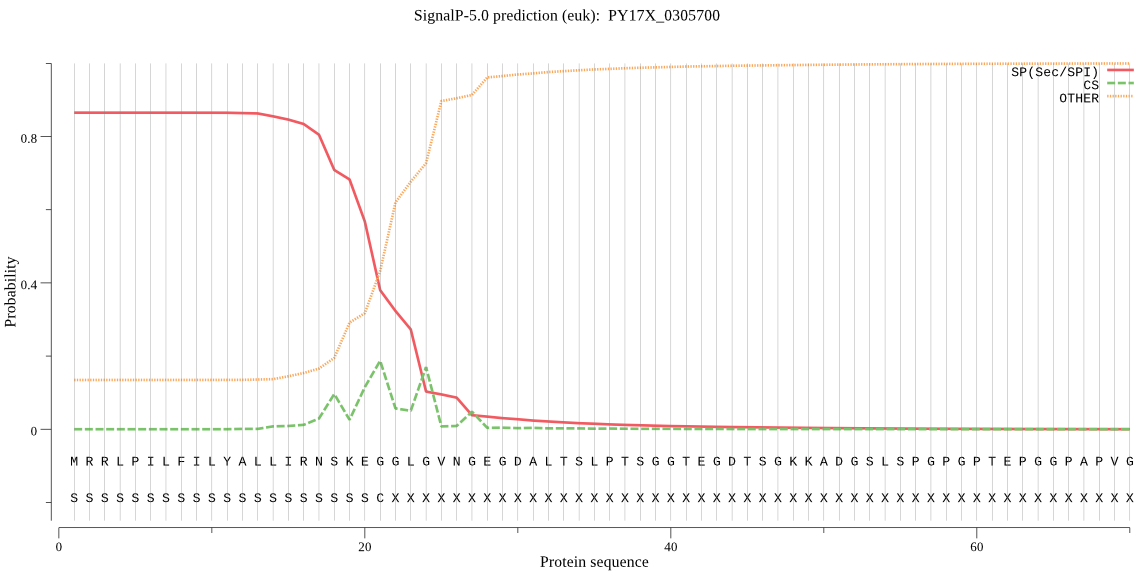
| PY17X_0305700 | SP | 0.045606 | 0.954275 | 0.000119 | CS pos: 24-25. GLG-VN. Pr: 0.1872 |
MRRLPILFILYALLIRNSKEGGLGVNGEGDALTSLPTSGGTEGDTSGKKADGSLSPGPGP TEPGGPAPVGPLPGSPQVEEPKLSTPSVEGSQLESSSLTKDGPKAVGEDGTLISGTEQEG SETDRTKQEKSGSDNNKQGGSPPGKPPGETITQDPSQQKIVDQNGPVKEEPGPKGPVLPP SSPGESPETKSELKTPELGTIGSGGLSPEPGKSNQSDTSIPTGSQSITADQITNTKGETG SPDSVQSIPESSGSSITDPKITKSEPGVSGPDGNLPESVKLDTPTLNGSQPSTDDQKVQA KGENELPNPVQSSSELGDPSKPGPKITNPGSGATGPDGTPSVPTKQGASAPTGPVPGNSD QGNVVSEPKESGDTEGKGNKGSNNPDDPNSKGKQSDNAPGSDNTLQTKKPGTDDASLENA KKLSQDTDNPPKVVEGTEHVEKPTISDSDPSAIKTEVTKGPQAPEIQLPSSEKSNTHTSD PNSDIHSQTLSPPVNNPISTDQDAVNTLESLQSDSPSAESVPSIIESIPFQDFDIKSAML KNYKGLKITGQCNSSFFMFFVPYVYIDVDTKINQISIFSTKKKVNGTTNDVPTQLIEFKS EDDTLQNQCADNKTFKIVIYLDNGVLHVKWKVYDPSSETEPNTNVDVRKYRLKNIETPIT SIQVHNVKRTLKNITIESKNYTINDDLPEKCEAIASDCFLHGNVDTEKCYKCALLYRDTP ITDKCYDYLPDDYKESLKTDRLVTAQSEDEINYELMGHIDNVLEGIYNIDEDNNNNNKEL KKFEELDDEIKKDMLLYCKELKESDVSGTLEEYALGDVEDIFYNLIQLINNNTGVVISKL KNKLMNPAICLKDVNQWGEKKKGLVLPELSSNNFDNLNDKNDDTTINSKLDEKLEEEGFD GVIDLPLPHENEFEGYTTIADFQYCNNEYCDRLKDNNSCISKIDVEEQGNCATSWLFASK FHLDTIGCMKGHENFSASALYVANCSKKDSKDKCLTGSNPLEFLNIIDENKFLPTTSNLP YSYKKVGEECPKAMDNWTNLWKDVKLLKHENNDKSLNANGYISYQSKDFKDNFSEYINLI KQEIQNKGSIIAYINSKDIISYDFNGSKIHKLCGSDTPDMAVNIIGYGKYINENNQVKSY WLVRNSWGKHWADEGNFKVDIETPENCEHNFIHTFATFNIYIPFVKKNIPSESEINLYYS KNSPDFYNNLYFKNLDATDTNKDSIIEGQNEPAKVADSEETNQLTGDGQLKSQQGGETKE LSGSDGEKKENEGVESETLPTDSEGETQKLKQEQQQQQQQQQQQLHSQPQPEPQPEPQPA PQVKVEGQEQVQEQVQEQRADSKQVDGAEITKNVTGQEEKKEIGTENSSSKGKEQQPAAK ASEGQQPVNVPEQQAGRAQAADSITKLQAVKIEFMHILKQIKNGRVQMNLVKYGDELNMI DDRMCSRVHAINYDKKDECEKFCITKWSECQTATSPGYCLSKLYESNDCYFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0305700 | 182 S | LPPSSPGES | 0.996 | unsp | PY17X_0305700 | 182 S | LPPSSPGES | 0.996 | unsp | PY17X_0305700 | 182 S | LPPSSPGES | 0.996 | unsp | PY17X_0305700 | 186 S | SPGESPETK | 0.991 | unsp | PY17X_0305700 | 207 S | SGGLSPEPG | 0.996 | unsp | PY17X_0305700 | 255 S | SSGSSITDP | 0.997 | unsp | PY17X_0305700 | 292 S | GSQPSTDDQ | 0.993 | unsp | PY17X_0305700 | 416 S | TDDASLENA | 0.993 | unsp | PY17X_0305700 | 446 S | KPTISDSDP | 0.991 | unsp | PY17X_0305700 | 471 S | QLPSSEKSN | 0.996 | unsp | PY17X_0305700 | 986 S | VANCSKKDS | 0.995 | unsp | PY17X_0305700 | 1262 S | TKELSGSDG | 0.997 | unsp | PY17X_0305700 | 1283 S | LPTDSEGET | 0.993 | unsp | PY17X_0305700 | 46 S | EGDTSGKKA | 0.997 | unsp | PY17X_0305700 | 131 S | KQEKSGSDN | 0.996 | unsp |