

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_0408000 | SP | 0.023919 | 0.975406 | 0.000675 | CS pos: 20-21. IEA-KP. Pr: 0.9647 |
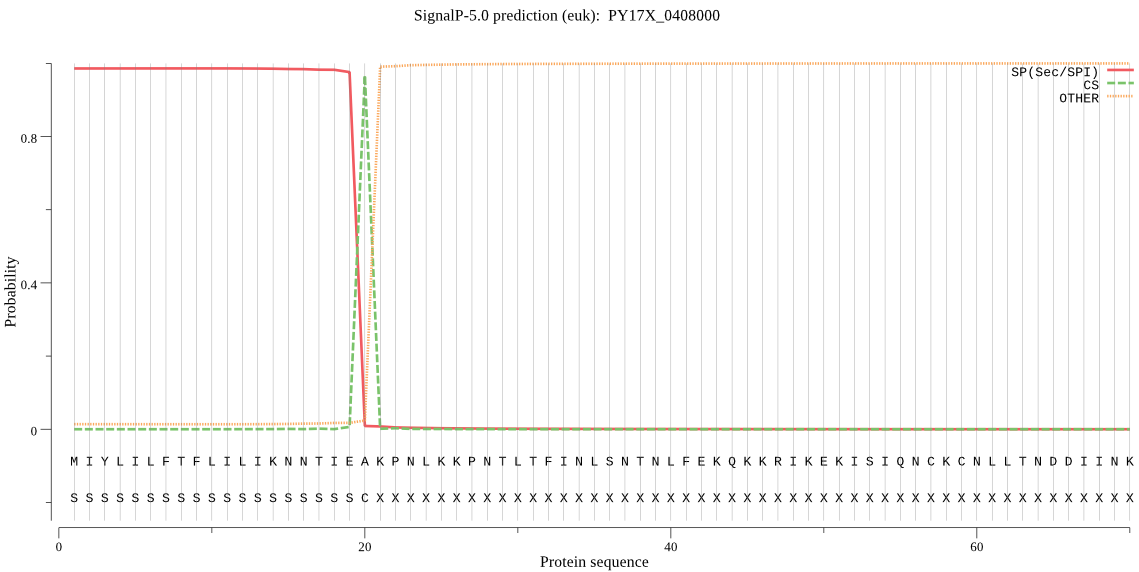
MIYLILFTFLILIKNNTIEAKPNLKKPNTLTFINLSNTNLFEKQKKRIKEKISIQNCKCN LLTNDDIINKNIEEDVNEINNVKNISENFDKTKVNINNDVTKIDQEEESKIRQNAMEKMV EKMVEKILKKKKYKKLEKLLCCNNSIKDMKKNIKLYFFKKRIIYLTDEINKKTSDELIKQ LLYLDNINHNDIKIYINSPGGSINEGLAILDIFNYIKSDIQTISFGLVASMASVILASGK KGKRKSFPNCRIMIHQPLGNAFGHPHDIEIQTKEILYLKKLLYYYLSKFTNQNEQTIEKN SDRDYYMNSFEAKKYGIIDQVIETKLPHPYFTDLTEN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0408000 | 174 S | NKKTSDELI | 0.994 | unsp | PY17X_0408000 | 301 S | IEKNSDRDY | 0.997 | unsp |