

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_0414900 | SP | 0.034900 | 0.964535 | 0.000564 | CS pos: 24-25. CDA-KV. Pr: 0.8358 |
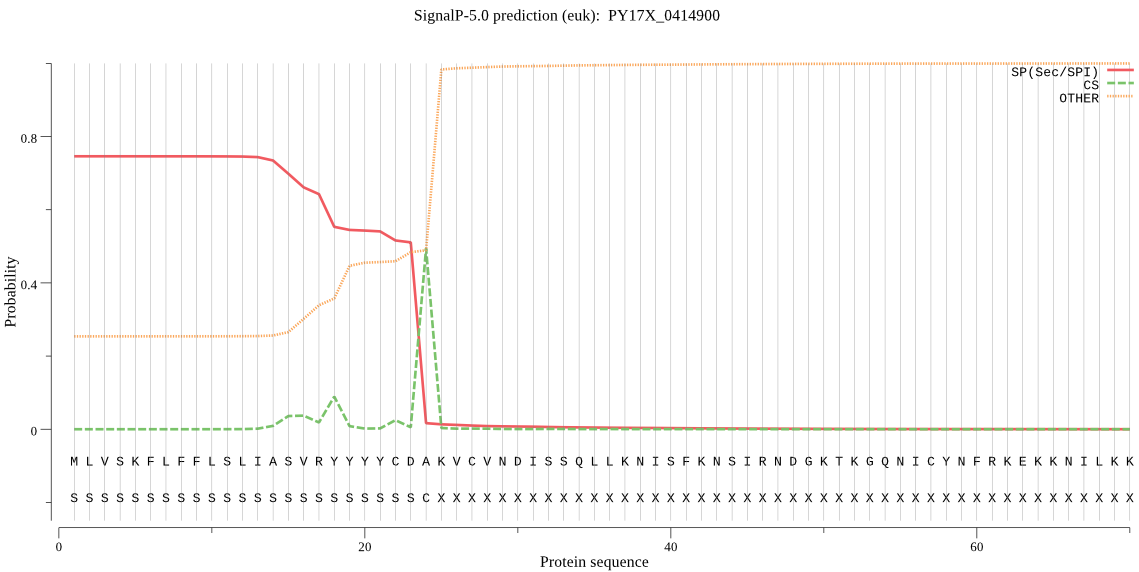
MLVSKFLFFLSLIASVRYYYYCDAKVCVNDISSQLLKNISFKNSIRNDGKTKGQNICYNF RKEKKNILKKKIKKKYSLNYSKEYTQKKKKKYNLYFLNSSKFTDIKENVKAWFNLDYIEN HYAYSYIKSFARIPPITKIYLLNTFILSVLIHLNKNVYKYILYDFDKIFKNGEIWRLVTP YFYIGNLYLQYILMFNYLNIYMSSVEIAHYKNPEDFLIFLTYGYISNILFTIIASMYNEN VMDLKKYIHKIINLIITIPSTNNKDDKKLVQKETYNHLGYVFSTYILYYWSRINEGSLIN CFDLFFIKAEYIPFFFVIQNILLYNEFSFFEVASILSSYTFFTYEKYFKSNFLRDLFSGF LKYIRVYPMYNMYKEEYE