

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_0704600 | OTHER | 0.999902 | 0.000001 | 0.000097 |
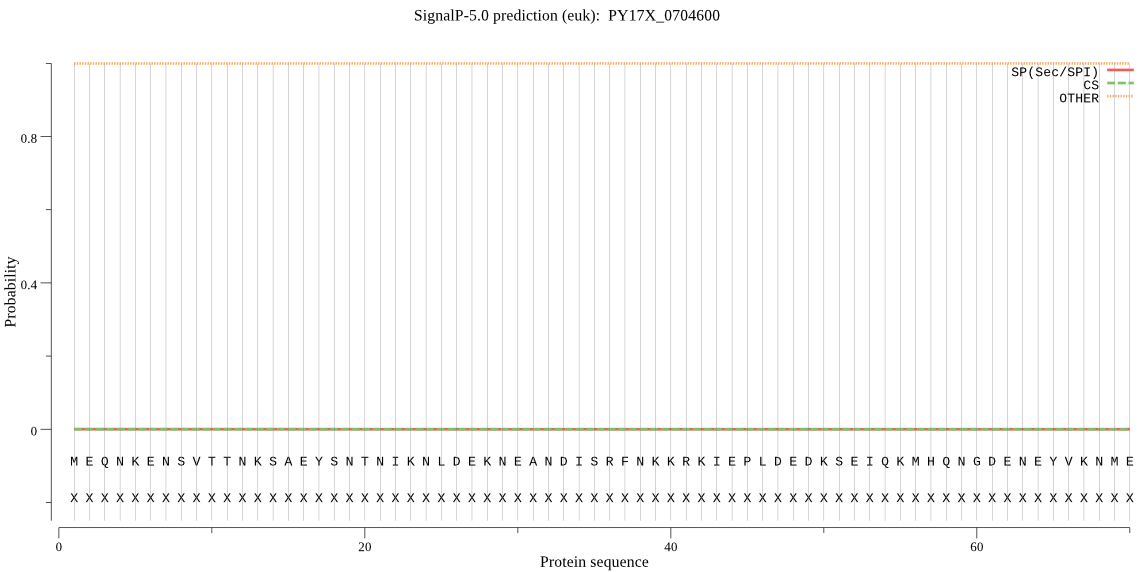
MEQNKENSVTTNKSAEYSNTNIKNLDEKNEANDISRFNKKRKIEPLDEDKSEIQKMHQNG DENEYVKNMEKDNKNQCLNNSYESGHEKNADKINKNNDKADKIGNNSNNEHTKRDNINVL EESKELKKENYKYYIDYRKYRNKNNVTFSTKYKDSCISLSSDKLTCYGDKGWSSVFVNNG ADVGKWYYEIKIEEPVHKSNFLGYKDTILKVNPYVRVGFACRYMRYDYPIGTDKYSYCVN SKNGKIFNNSISYDCMEPIKVGDIIGCYINLKNKNSYTFDPRSDKKLYEYLQNGILCDPK NPPILKKNYGSFIFFSLNGQIKKNAFIDIYEGFYHPSVSLYMGASAKINLGPKFTYRHLN DYVPCIYMDPPIIL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_0704600 | 283 S | FDPRSDKKL | 0.99 | unsp |