-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
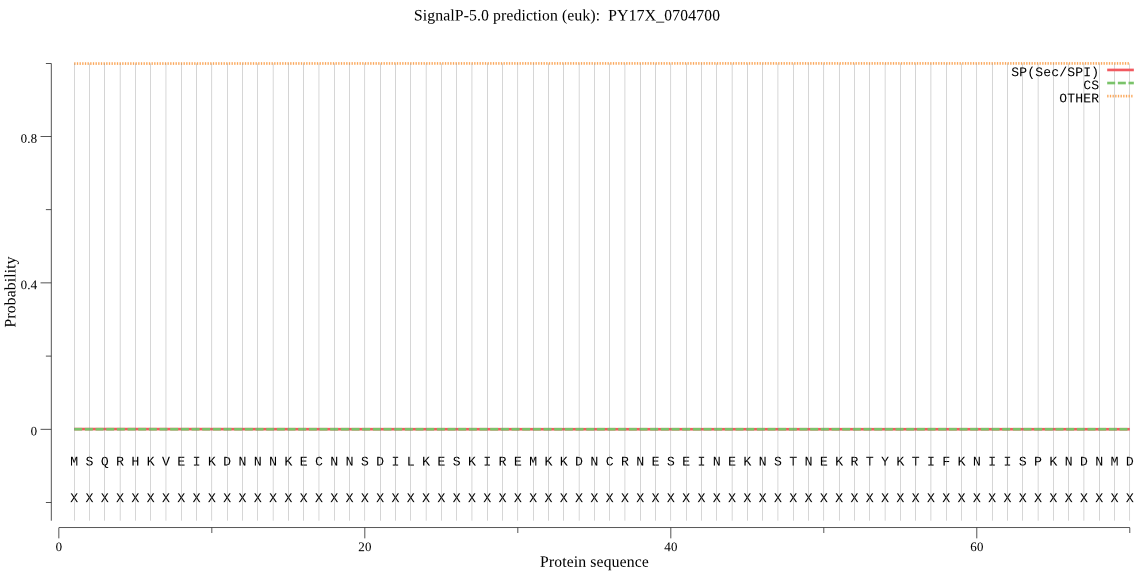
PY17X_0704700 | OTHER | 0.999998 | 0.000000 | 0.000002 | |
No Results
-
Fasta :-
>PY17X_0704700
MSQRHKVEIKDNNNKECNNSDILKESKIREMKKDNCRNESEINEKNSTNEKRTYKTIFKN
IISPKNDNMDYDVKDWHDKKKKKKKKKKSKNENEIAKDEKDNNNNNDDDDEKFITYKIPK
YLRKRNIKCPFIYKQVFYGKYGIVNYDLQGSNKETLVITFHGLNGTNLTFLEIQKILIRY
KFQVLNFDLYGHGLSACPKYNHRNKTYGIDFFLSQTEELLTHLNLLDRNFYLIGFSMGCI
IATNFAKKYIKQVKKIILISPVGALEKKPFLVKLLKIFPCIINMSSFFMLPYLISKRNFK
KKKTKISNDMDSEDEASYYMYNRIMWQAFAKKNITHSILGCINNLKMWSSHEIFKEVGLH
HIPVLILCGDKDNICSVHVLKNTSKLFANSHLIIFKNASHLVLVDKSREINSCVLIFFLS
SNNTNLKACQHMFPVDNIET
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PY17X_0704700.fa
Sequence name : PY17X_0704700
Sequence length : 440
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.236
CoefTot : -0.260
ChDiff : 27
ZoneTo : 7
KR : 2
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.771 1.706 0.296 0.664
MesoH : -0.042 0.854 -0.255 0.344
MuHd_075 : 9.659 5.398 5.025 2.152
MuHd_095 : 23.599 9.561 5.712 3.871
MuHd_100 : 19.949 7.613 5.204 3.373
MuHd_105 : 12.145 5.973 4.123 2.068
Hmax_075 : -16.700 -5.775 -5.824 -1.811
Hmax_095 : -10.238 -4.900 -5.086 -0.481
Hmax_100 : -13.700 -7.900 -6.957 -0.800
Hmax_105 : -17.700 -6.400 -6.365 -1.560
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9483 0.0517
DFMC : 0.8960 0.1040
-
Fasta :-
>PY17X_0704700
ATGTCACAACGACACAAAGTTGAAATAAAAGACAATAATAACAAAGAGTGCAATAATAGT
GATATATTAAAAGAATCAAAGATCCGAGAAATGAAGAAAGACAATTGTAGAAATGAAAGT
GAAATAAACGAAAAGAATAGCACAAATGAAAAACGAACTTACAAAACAATTTTTAAAAAT
ATAATTTCACCAAAAAATGATAATATGGACTATGATGTAAAAGATTGGCATGATAAAAAA
AAAAAAAAAAAAAAAAAAAAAAAATCGAAGAATGAGAATGAAATAGCGAAAGATGAAAAA
GATAATAATAATAATAATGATGATGATGATGAAAAGTTTATTACATACAAAATACCAAAA
TATTTAAGGAAAAGAAATATCAAATGCCCATTTATATATAAACAAGTATTTTATGGAAAA
TATGGTATAGTAAACTATGATCTACAAGGAAGTAATAAAGAAACTTTAGTTATAACTTTT
CATGGATTAAATGGTACTAACTTAACATTTTTAGAAATTCAAAAAATATTAATTCGATAT
AAATTTCAAGTTCTTAATTTTGATTTATATGGGCATGGATTATCTGCATGCCCAAAATAT
AATCATAGAAATAAAACATATGGTATTGATTTTTTTTTATCTCAAACTGAAGAATTATTA
ACTCATTTAAATTTATTAGACAGAAATTTTTATTTGATTGGATTTTCTATGGGTTGTATT
ATTGCTACTAACTTTGCAAAAAAATATATTAAGCAAGTAAAAAAAATTATTTTAATATCA
CCTGTTGGGGCTTTAGAAAAAAAACCATTTTTAGTAAAACTATTAAAAATTTTCCCATGC
ATAATTAACATGTCCTCTTTCTTTATGTTACCATATCTTATTTCAAAACGAAATTTCAAA
AAAAAAAAAACGAAGATTTCCAATGATATGGATTCAGAAGATGAAGCATCCTATTATATG
TATAATCGAATAATGTGGCAAGCTTTTGCTAAAAAAAATATTACTCATTCTATTTTAGGA
TGTATTAATAACTTAAAAATGTGGAGTTCCCATGAAATATTTAAAGAAGTTGGCCTACAT
CATATACCCGTTTTAATTTTATGTGGAGATAAAGATAATATATGTAGTGTACATGTACTT
AAAAATACATCTAAGCTTTTTGCTAATTCTCATCTAATCATATTTAAAAATGCTTCGCAT
CTAGTTCTTGTAGATAAAAGCAGAGAAATTAATTCATGTGTTCTTATATTTTTCCTATCC
TCAAATAATACAAATTTGAAAGCATGCCAACATATGTTTCCAGTTGATAATATAGAAACA
TAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
PY17X_0704700 | 151 S | DLQGSNKET | 0.991 | unsp | PY17X_0704700 | 151 S | DLQGSNKET | 0.991 | unsp | PY17X_0704700 | 151 S | DLQGSNKET | 0.991 | unsp | PY17X_0704700 | 312 S | NDMDSEDEA | 0.994 | unsp | PY17X_0704700 | 47 S | NEKNSTNEK | 0.993 | unsp | PY17X_0704700 | 89 S | KKKKSKNEN | 0.997 | unsp |


