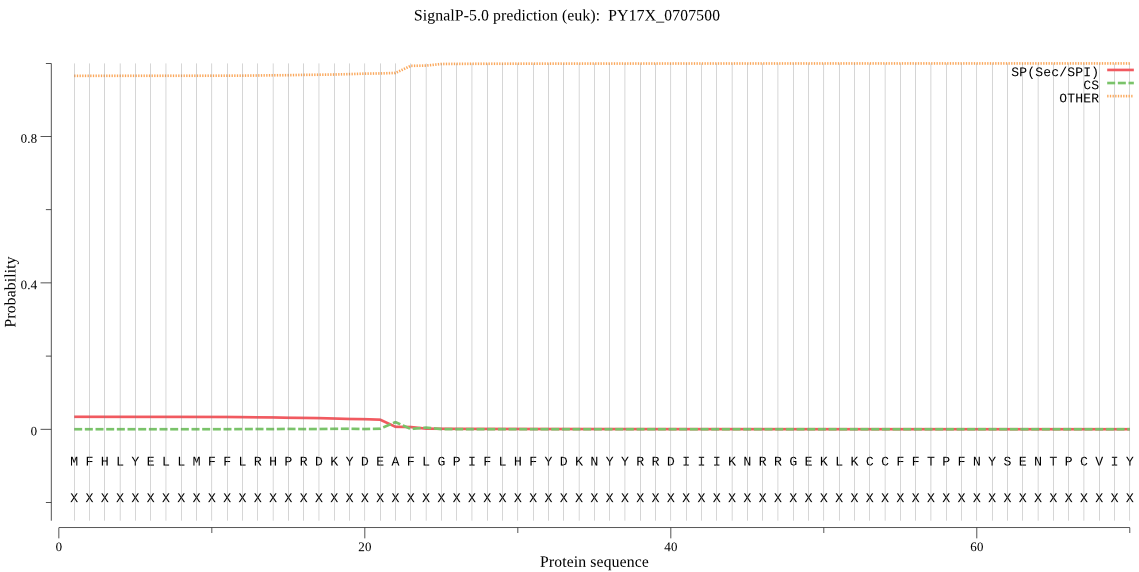
Fasta :-
MFHLYELLMFFLRHPRDKYDEAFLGPIFLHFYDKNYYRRDIIIKNRRGEKLKCCFFTPFN
YSENTPCVIYTHSTSSCQLEVLDILHILLVCECSVFSFDCAGCGLSDGYYSTKGWNETQD
LYLILNHLRNVEKIKNFVLWGKHSGAASSIIAAALDQNIKMLILESPFVSLIELYKTTFN
LCAKKKKEIIFKNICLYFTRRKIKKKFNYDINNVCPVFFIEDITIPTIYIISKSDSIVHP
AHSLYLAYKQKKAKKIIYIAEKGTQSHEAYTYDSKLIIAMKSILYSYNFENDIRDKIFDV
STYYKLFTFLREKYAYEFDYIDRLITKKNNKKNKIIDKVKKFVCFKYQSKLSLSSSTGLN
QSSVDSLRTSKVYENECTFQTGENNNDPIQVCDQINGSGCSNDYQDVFSDEDNMLDNPTE
NNNMNTDYNIDKFNKLNEHNNKFEEIQGTSEKPFKLLHMQSSRSSLKSNKNTYKKSLTWN
SNLQSSITYFKEDCPLSLQKN