

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
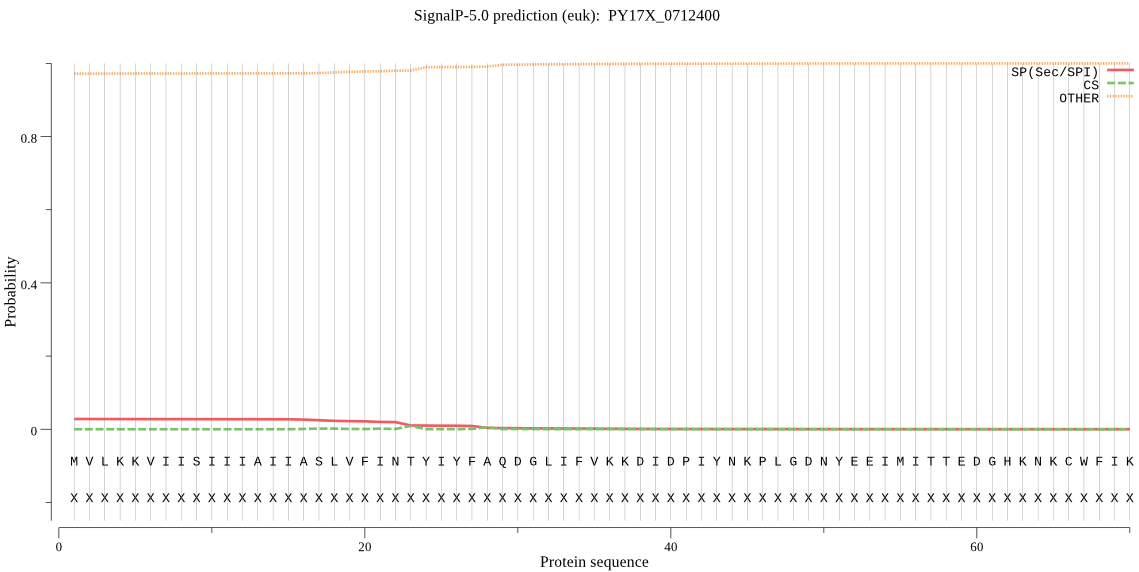
| PY17X_0712400 | OTHER | 0.857883 | 0.141146 | 0.000972 |
MVLKKVIISIIIAIIASLVFINTYIYFAQDGLIFVKKDIDPIYNKPLGDNYEEIMITTED GHKNKCWFIKTQDYENKPVILYFLGNGSYVEKNVEIFNLMVGRVDVSIFSCSHRGTGDNT LSPSEASFYSDAQTYLNYLKMKNMKNIFLFGTSMGCAVAIETALNNSNSVAGLIVQNPFL SMKKMAKLAKPFLFFIILSYDLLIRTKMDNEEKIKKNRVPVLFNISEKDKIVPPDHGKKL YEICPSQKFIYTAKDGEHNNILVNDDGSYHRSMKIFIETVNSTYSKK
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_0712400 | 122 S | DNTLSPSEA | 0.993 | unsp |