

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
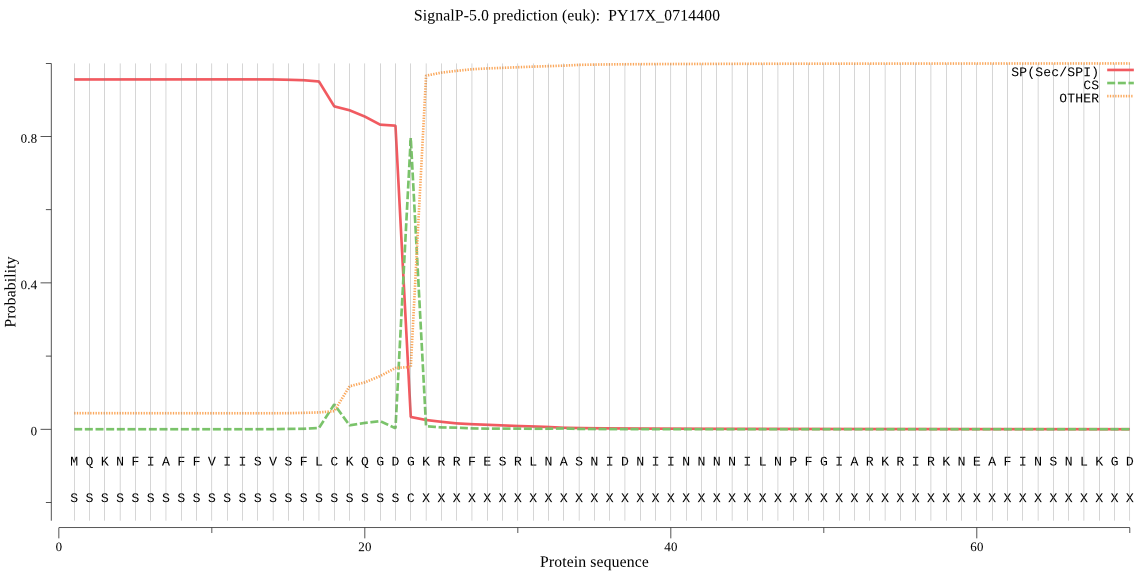
| PY17X_0714400 | SP | 0.007823 | 0.991330 | 0.000847 | CS pos: 23-24. GDG-KR. Pr: 0.9530 |
MQKNFIAFFVIISVSFLCKQGDGKRRFESRLNASNIDNIINNNNILNPFGIARKRIRKNE AFINSNLKGDDILKRDKKKGFKSKINENKDEYNIKSKKYDQSGIKNRLNYSTNDQDNVNN YFFNKNENNFINKRNKMNQLFMSDEEYTINSDDYTEKAWDAISSLNKIGEKYESAYVEAE MLLLALLNDSPDGLAQRILKEAGIDTDLLSHDIDVYLKKQPKMPSGFGEQKILGRTLQTV LTTSKRLKREFNDEYISIEHLLLSIISEDSKFTRPWLLKYNVNYEKVKKAVEKIRGKKKV TSKTPEMTYQALEKYSRDLTALARAGKLDPVIGRDEEIRRAIQILSRRTKNNPILLGDPG VGKTAIVEGLAIKIVQGDVPDSLKGRKLVSLDMSSLIAGAKYRGDFEERLKSILKEIQDS EGQVVMFIDEIHTVVGAGAVAEGALDAGNILKPLLARGELRCIGATTVSEYRQFIEKDKA LERRFQQILVDQPSVDETISILRGLKERYEVHHGVRILDSALVQAAILSDRYISYRFLPD KAIDLIDEAASNLKIQLSSKPIQLDNIEKQLIQLEMEKISILGDKQTASLINKSSSGNDD NNVSTDYTQSQNFIKKRISEKEINRLKTIDHIMNELRKEQKNILESWTSEKMYVDNIRAI KERIDVVKIEIEKAERYFDLNRAAELRFETLPDLEKQLKNAEENYVNDIPERNRMLKDEV TSEDIMNIVSISTGIRLNKLLKSEKEKILNLENELHKQIIGQDDAVKIVARAVQRSRVGM NNPKRPIASLMFLGPTGVGKTELSKVLADVLFDTPDAVIHFDMSEYMEKHSISKLIGAAP GYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRVIDEGKLSDTKGNVANFRN TIIIFTSNLGSQSILELANDPNKKEKIKEQVMKSVRETFRPEFYNRIDDHVIFDSLTKKE LKEIANIEITKVANRLFDKNFKISIDDAVFSYIVDKAYDPSFGARPLKRVIQSEIETEIA IRILNETFVENDTIRVSLKDQKLHFSKG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0714400 | 596 S | NKSSSGNDD | 0.994 | unsp | PY17X_0714400 | 596 S | NKSSSGNDD | 0.994 | unsp | PY17X_0714400 | 596 S | NKSSSGNDD | 0.994 | unsp | PY17X_0714400 | 619 S | KKRISEKEI | 0.998 | unsp | PY17X_0714400 | 743 S | KLLKSEKEK | 0.993 | unsp | PY17X_0714400 | 934 S | QVMKSVRET | 0.997 | unsp | PY17X_0714400 | 984 S | NFKISIDDA | 0.993 | unsp | PY17X_0714400 | 1013 S | RVIQSEIET | 0.991 | unsp | PY17X_0714400 | 1037 S | TIRVSLKDQ | 0.998 | unsp | PY17X_0714400 | 382 S | DVPDSLKGR | 0.991 | unsp | PY17X_0714400 | 534 S | DRYISYRFL | 0.993 | unsp |