

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
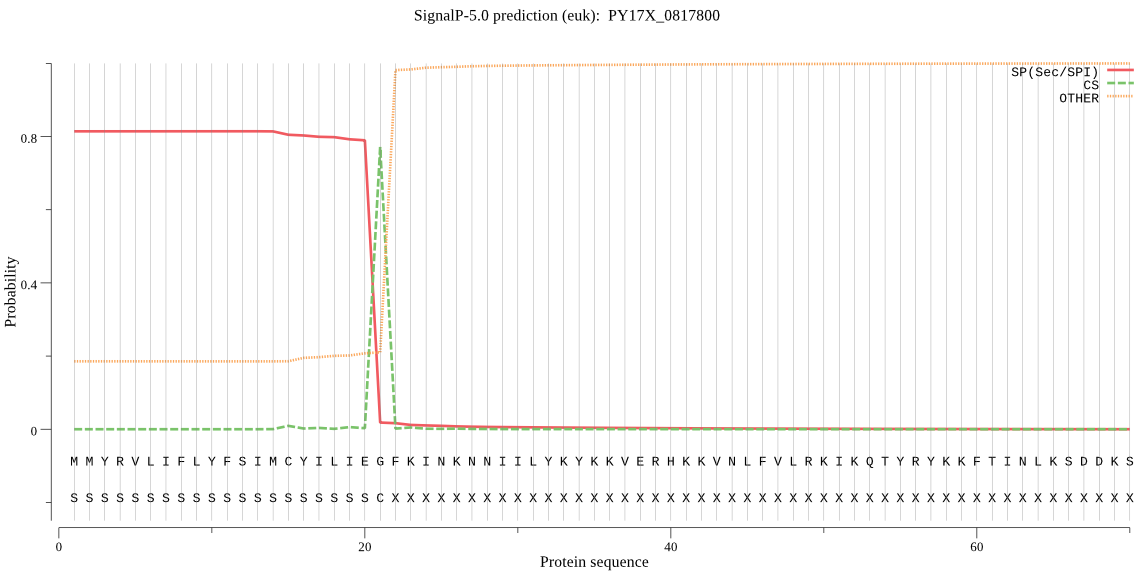
| PY17X_0817800 | SP | 0.496098 | 0.503182 | 0.000720 | CS pos: 21-22. IEG-FK. Pr: 0.9583 |
MMYRVLIFLYFSIMCYILIEGFKINKNNIILYKYKKVERHKKVNLFVLRKIKQTYRYKKF TINLKSDDKSLNNIKEKKNIIKENIFNFYKKNIYLFSKILLYIKNGTNFITEKFPISVIY QIFISISFFFLHFYLLSKNFIILLPYQLIPNYSNNLMSLDLNSAFFIFSSLFFLKNFISN FQKFKTRLLSNKFTIPLKETKKKNILTISVLFLASYILSGYVSIYAEQILYYSKSFNFRI SDATIKSLQILIGHLMWVGCSIQIFRTFLYPYFKNNVSNLNFRYKDSWCFKVIYGYVCSH YIFNLIDLLNNTIINYLHNDDEIYIENNIDNIINEKKFLSTFICIISPCFSAPFFEEFIY RFFVLKSLCLFMNIHYAVIFSSLFFAIHHLNIFNIIPLFFLSFFWSYIYIYTDNILVTML IHSFWNIYVFLSSLYN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_0817800 | 190 S | TRLLSNKFT | 0.994 | unsp |