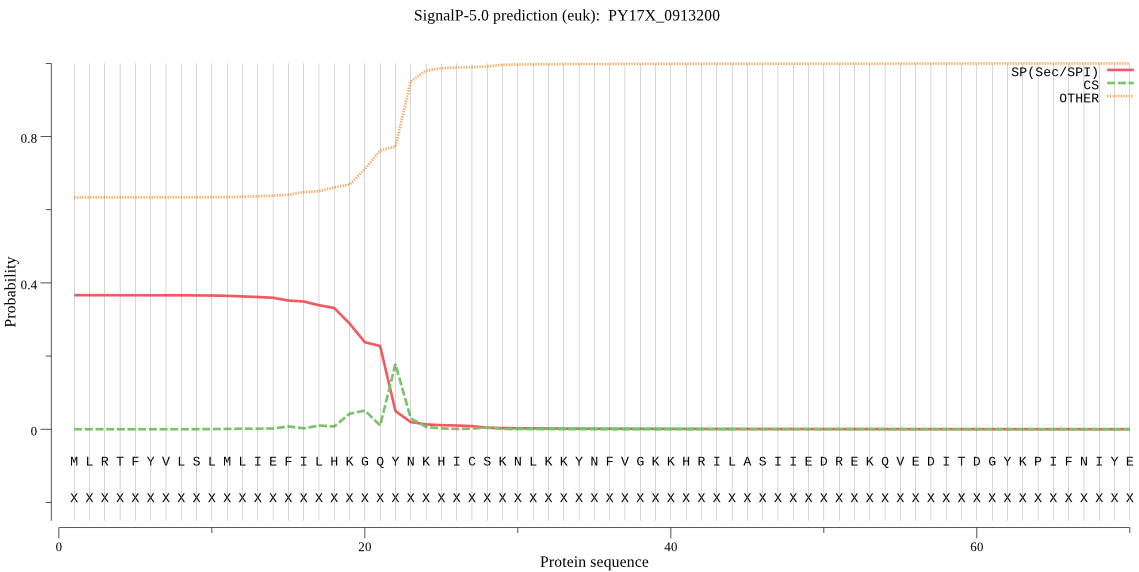
Fasta :-
MLRTFYVLSLMLIEFILHKGQYNKHICSKNLKKYNFVGKKHRILASIIEDREKQVEDITD
GYKPIFNIYEISAAFHKKKDIADKKKRRRYGNQQSIENRRIAEENERRLSNQLDDIQFIE
LSNKYPNIGKQNSQQNKVNKINNQNGASNSNDNIRNDEDEDEGEDEDEDDDLIEGRKDNL
EEDDLVEKNGANLKRGNMHGQEEKNKNINTTPGNENNSKNVNDNKKSGISLKDKIDNNEQ
HNSGLKGTTKYLDDNIKTYTFDHYKLITNSDNILNDIKVDASDISKLSINSLSIEYNEVN
KTEYTHQRHIVLTNKGNRRYKIFLMTKNPKFTKTEDIEEPEMSFIQTETGENTNEKEDEE
NYLNENLYSGFGTIDYENGYSKKKKKINSEHASELNDKISNSQNIEKSDSHENEKYNHGF
IGKIQSFFSFLSIPSSKKDDSIGSEKKSEERNNVDSKPKLNKKPNDTAKKNNSNKILTVD
KVTDQYLLNLKNKNMKEQELIFIFHGDLDLHSKKMKTIINEANVKFTKYINMHFKDVKNI
RYDISSPINFVCFFIPIIFDMSNLKILKEALIILHNELKNYIDNWNFSNTYIAFDTDYEN
EDIDNAMNKLNENMKKYIKKPKKLYNIKYSFLRKMWGLESIFSLSKNRNQKNAGIEEKIL
NALPKELKEYSTWNLSFIRVFNAWLLSGYGNKNVKICVIDSGVDKNHIDLAKNIYTPKYS
DRYEMTDDFFDFMVKNPIDTSGHGTHVSGIAAASANSLGMVGVAPDVNLISLRFIDGDSY
GGSFHVIKAINVCILNKSPIINASWGSRNYDTNMFLAIERLKYTFKGKGTVFIAAAGNEN
KNNDLYPIYPASYKLPNVYSVGSINKFLQISPFSNYGANSVHILAPGHHIYSTTPMNTYK
MNTGTSMAAPHVSGVAGLIYSVCHKQGFIPESDEVLDIITRTSIKIVSKDKKTIHNSLIN
AEAAVLTTLLGGLWMQMDCHFAKFYLNKDQQKNIPVVFSAYKDGMYESDIIIGIQPEDSN
SKEYGEIVIPIKILTNPKLKNFNLSPRVGKKIHIDANESNDDILSYICENALYNLYEHDN
SFLISSLILFFIGIILIALASIVFFLKHHQSKQRDAEKYMHQKMVDRAHGIKYNFKDAGA
DGIKRINTMDDNINNHRNTQRFTIVQNEDNMYVLKKKSSIQAKYEPRNELVKRPLVKRPI
VKHADINVNFKNIDELYEPQNNSPE