

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
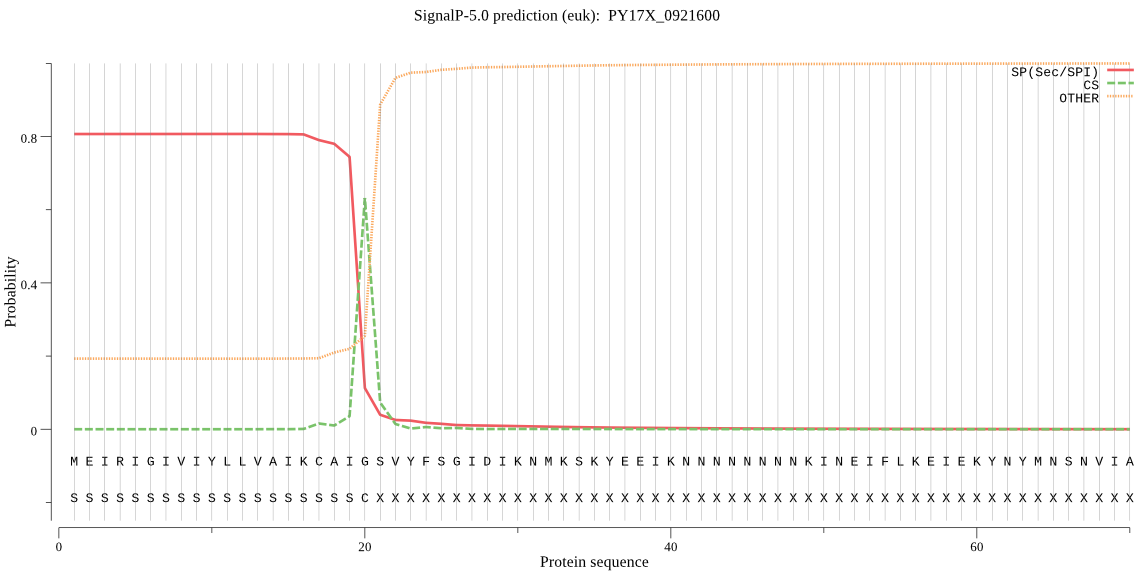
| PY17X_0921600 | SP | 0.023406 | 0.976178 | 0.000416 | CS pos: 20-21. AIG-SV. Pr: 0.9323 |
MEIRIGIVIYLLVAIKCAIGSVYFSGIDIKNMKSKYEEIKNNNNNNNNKINEIFLKEIEK YNYMNSNVIALSTSRHYFNYRHTANLLIAYKYLKKNGDIMDKNILLMIPFDQACNCRNII EGTIFKNYEKLPNEYINKNMKENLYNKLNIDYKNDNINDEQLKKVIRHRNNSFTPYNNRL YTNEYKKKNLFIYITGHGGINFLKIQEFSIISSSEFNLYIQELLIKKIYKNIFVIIDTCQ GYSFYDDILRFINKNKIKNVFLLSSSNRHENSYSFFSSKYLSVSTVDRFTYNFFDYMENI HKMESKEIYKNLKFFSIQNILNYLKTKNLMSHPSINNSKLNVSSFLHDQNVIFYDSTMLF PKLFYNKLNKNDKNDYPNNNHDHIHSEHTCFGYLGICGHIKSQIYLNMESLYNDRSYYNN DDVYNSYEFYFTDKYFYFKYFDIYYMGIFLILLFVILLFLIVYMLY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0921600 | 25 S | SVYFSGIDI | 0.995 | unsp | PY17X_0921600 | 34 S | KNMKSKYEE | 0.991 | unsp |