

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
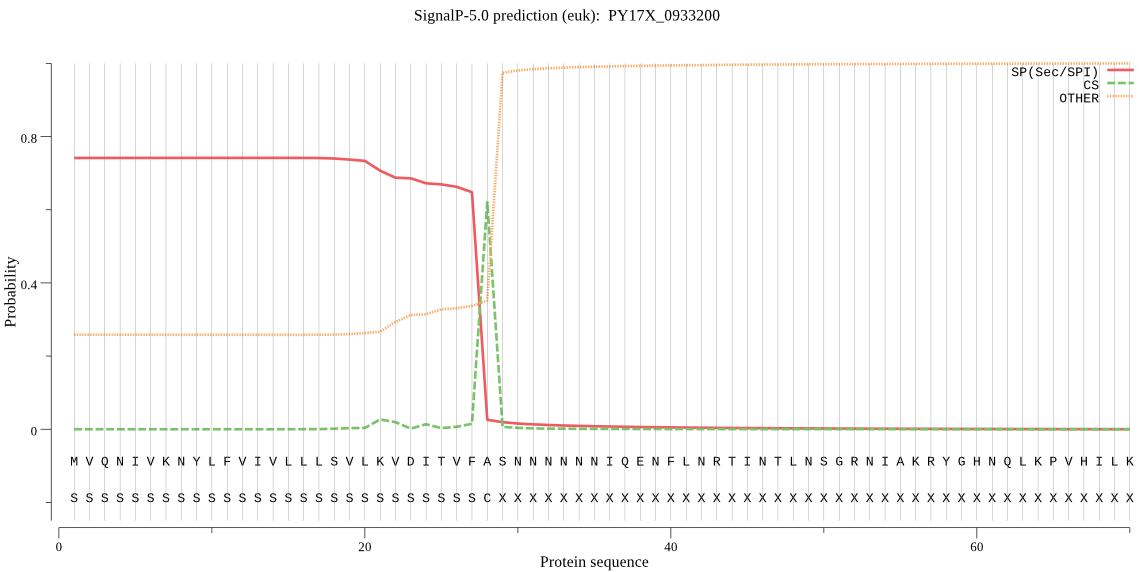
| PY17X_0933200 | SP | 0.151790 | 0.832235 | 0.015976 | CS pos: 28-29. VFA-SN. Pr: 0.8416 |
MVQNIVKNYLFVIVLLLSVLKVDITVFASNNNNNNIQENFLNRTINTLNSGRNIAKRYGH NQLKPVHILKSIIKSEYGLNLFKSNNIDLGNLKEYTDAALEQTRAGAPLDNKTIVINSEA TNEVLAEAKAIAKKYKSPKVDIEHILYGLMSDELVSEIFGEIYLTEDSIKEILKNKFEKA TKTKEKKTTGLNIEQFGSNLNEKVRSGKLQGIYGRDEEIRAIIESLLRYNKNSPVLVGQP GTGKTTIVEGLAYRIEKGDVPKELLGYTIISLNFRKFTAGTSYRGEFENRMKNIIKELKN KKNKIILFVDEIHLLLGAGKAEGGVDAANLLKPVLSKGEIKLIGATTVTEYRKYIESCSA FERRFEKVIVEPPSVETAIKILRSLKSKYEKFYGINITDKALVAAVKVSDKFIKDRFLPD KAIDLLNKACSFLQVQLSGKPRVIDLTERYIERLAYEISTLERDVDKVSKKKYSSLVEEF ELKKEELKKYYEEYVTTGERLKRKKEVEKKLNELKDIAQNYINAGQEPPTELQESLNEAQ KQFVEIYKNTVAYVEEKTHNAMNVDSLYQEHVSYIYLRDSGMPLGSLSFESSKGALKLYN SLSQSIIGNEDIIKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFN SKNNLIRVNMSEFTEAHSVSKITGSPPGYVGFSDSGQLTEAVRERPHSVVLFDELEKAHP DVFKVLLQILGDGYINDNHRRNIDFSNTIIIMTSNLGADLFKKQFFFDADNSNTQEYKRL FEDLRIQLIKKCKKVFKPEFVNRIDKIGIFEPLSKKNLHKIVKLRFKKLEKRLEEKNISI DVSDRAIDYIIDQSYDPELGARPTLIFIESVIMTKFAVMYLEKELIDDMDVYVDYNKTIN NIVINLSLS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0933200 | 708 S | ERPHSVVLF | 0.994 | unsp | PY17X_0933200 | 708 S | ERPHSVVLF | 0.994 | unsp | PY17X_0933200 | 708 S | ERPHSVVLF | 0.994 | unsp | PY17X_0933200 | 137 S | KKYKSPKVD | 0.997 | unsp | PY17X_0933200 | 475 S | KKYSSLVEE | 0.995 | unsp |