

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_0933300 | SP | 0.001569 | 0.998422 | 0.000010 | CS pos: 25-26. VSA-DL. Pr: 0.9680 |
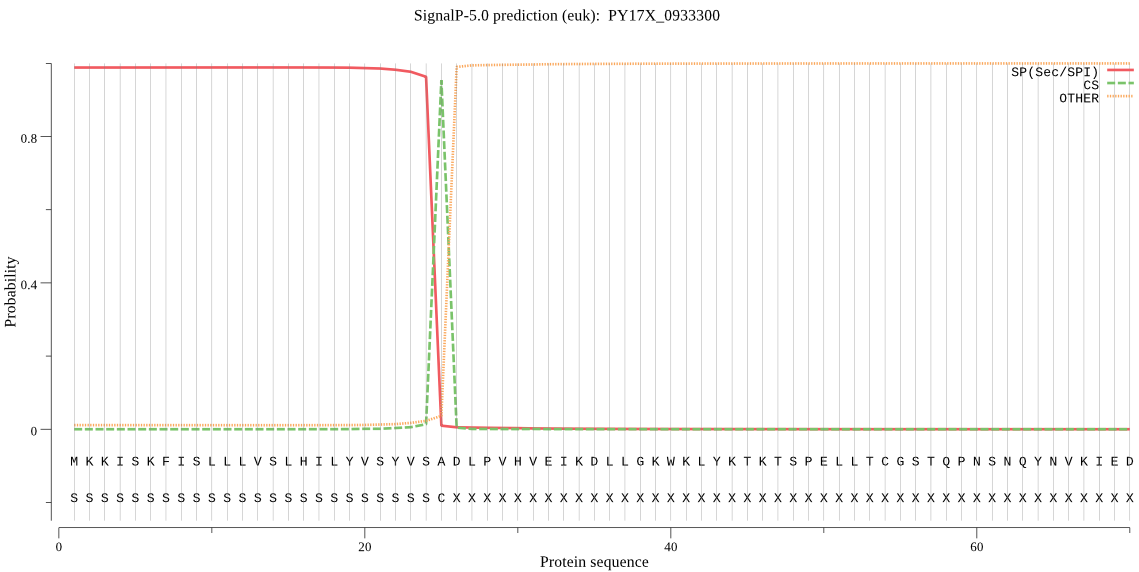
MKKISKFISLLLVSLHILYVSYVSADLPVHVEIKDLLGKWKLYKTKTSPELLTCGSTQPN SNQYNVKIEDYKKYLIDNHYPFDSELNVVLSNDFVKYGDVHDITGNEHRKNWNVLAVYDE KRRKKIGTWTTIFDQGFEIRIDNETYTAFMHYEPTGKCPEPSDEDITDSNGETICYSTSY DKTRFGWIDIMNKNNEQLHGCFYAEKYDTINDSNNYKIILNRSISGKKEPVITENKTCTS NQITDFDSYEPKTYIKANKVKLNKNSEMYWHKMKHDGKKKPLPEYMLKVQNQKYACPCNP NENIDNERSDVDPDSPVSPNMIELGNSNVDTNELDLNAYEEIKKSKHTELELNEMPKNFT WGDPFNNNVREYEVIDQLTCGSCYIASQMYVIKRRIEIGLTKLLETKYANDFDDALSLQT VLSCSFYDQGCHGGYPFLVSKMAKLHGIPLNSEFPYTAKQSTCPYPVNKGIPLSMIEAGS INKTESVPKDIKPSFRETIASALAGNDTKDNNNNVIDSGDPNRWYIKEYNYVGGCYGCNQ CDGEKIIMNEIYRNGPVVGSIEVTPNFYNYVDGVYYDKGFPHAKKCTVDVHKDNGYVYNI TGWEKVNHAIVILGWGEETIDGKLYKYWICRNSWGNGWGKEGYFKMIRGVNHVAIENHAI YIDPDFTRGAGKVLLEKMKNH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_0933300 | 309 S | DNERSDVDP | 0.994 | unsp | PY17X_0933300 | 309 S | DNERSDVDP | 0.994 | unsp | PY17X_0933300 | 309 S | DNERSDVDP | 0.994 | unsp | PY17X_0933300 | 494 S | DIKPSFRET | 0.995 | unsp | PY17X_0933300 | 162 S | CPEPSDEDI | 0.995 | unsp | PY17X_0933300 | 225 S | NRSISGKKE | 0.995 | unsp |