

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
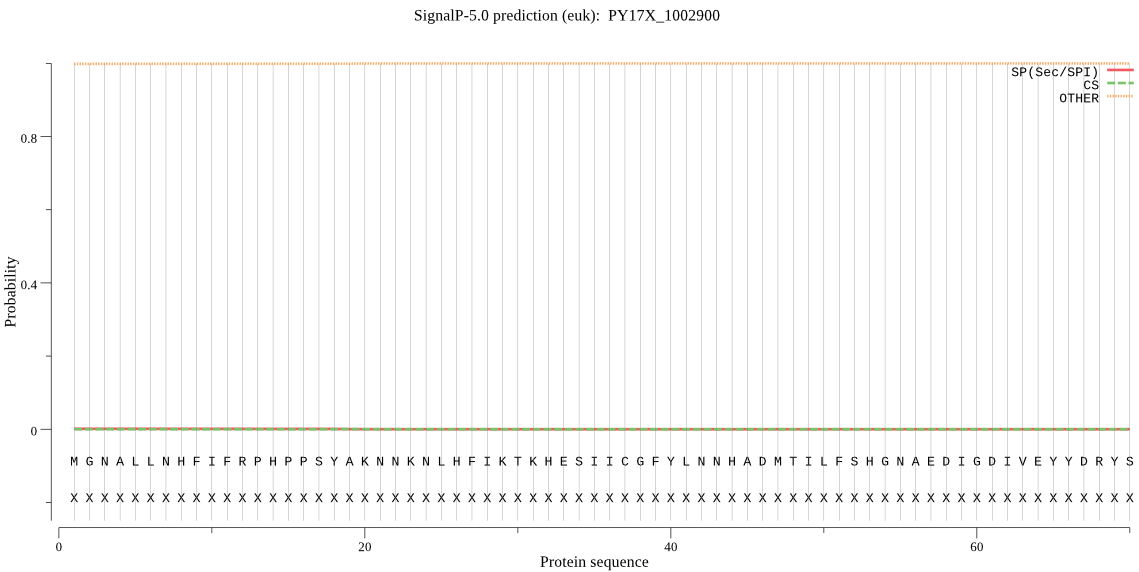
| PY17X_1002900 | OTHER | 0.991152 | 0.000736 | 0.008112 |
MGNALLNHFIFRPHPPSYAKNNKNLHFIKTKHESIICGFYLNNHADMTILFSHGNAEDIG DIVEYYDRYSKYIKVNMFMYDYSGYGHSTGYPNEEHIYNDVEAVYDYMITSLSIPSEKII AYGRSLGSTASVHIATKKNIKGLILQCPIASIHRVMFRLKHTLPYDLFCNIDKIHNVNCP ILFIHGMKDRVISYHGTMDMLKRVKVNTYYTFIEEADHNDIERFYFKELNSSIVTFMYIL KTNTRYIDNIVNDISKLSIHKLRNKYMLNNNDGVKTKLKGIMSEIQKSNHKTKPENKRNN TFYRVNLNPSQTSNNENDYNTVKENQESSKDVTKISNENDHYSSVTSISTIFEPEEWKTE GIPNKTFYESDDMNLEDISEYFYNNVSLLHLYKAKKWEKIINENYNKNSKLQFCKNKIND TSQKDICRKYYTENSNNNNSNECTKYRNINQNTNNNNFIRVSNTSINGSNTNKSGSNTSK SGSNTSKSGSNTSKSGSNTSKSGSNTSRSESKIRNNNGENKIIYGASGNLGVNSNSIASK KGANSGSSSYIYNPKRSINAKSNLKNYNTQSSEKLISNGVSNSISSIKRDELSSREGIAN FYNIKSKSNISSNMSVTSDKKIIAKGKQSRNNSHEKGGSNSSGNISGNGASNDNLKCCLS DNVNNNKIEINKLKGKIQVNNNNVSKYYETSNNETNFFDISQENNTLESSNKKQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1002900 | 474 S | NTNKSGSNT | 0.99 | unsp | PY17X_1002900 | 474 S | NTNKSGSNT | 0.99 | unsp | PY17X_1002900 | 474 S | NTNKSGSNT | 0.99 | unsp | PY17X_1002900 | 481 S | NTSKSGSNT | 0.994 | unsp | PY17X_1002900 | 488 S | NTSKSGSNT | 0.994 | unsp | PY17X_1002900 | 495 S | NTSKSGSNT | 0.994 | unsp | PY17X_1002900 | 502 S | NTSKSGSNT | 0.994 | unsp | PY17X_1002900 | 507 S | GSNTSRSES | 0.995 | unsp | PY17X_1002900 | 511 S | SRSESKIRN | 0.996 | unsp | PY17X_1002900 | 586 S | NSISSIKRD | 0.996 | unsp | PY17X_1002900 | 593 S | RDELSSREG | 0.996 | unsp | PY17X_1002900 | 615 S | SSNMSVTSD | 0.99 | unsp | PY17X_1002900 | 633 S | SRNNSHEKG | 0.993 | unsp | PY17X_1002900 | 328 S | ENQESSKDV | 0.996 | unsp | PY17X_1002900 | 422 S | INDTSQKDI | 0.997 | unsp |