

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
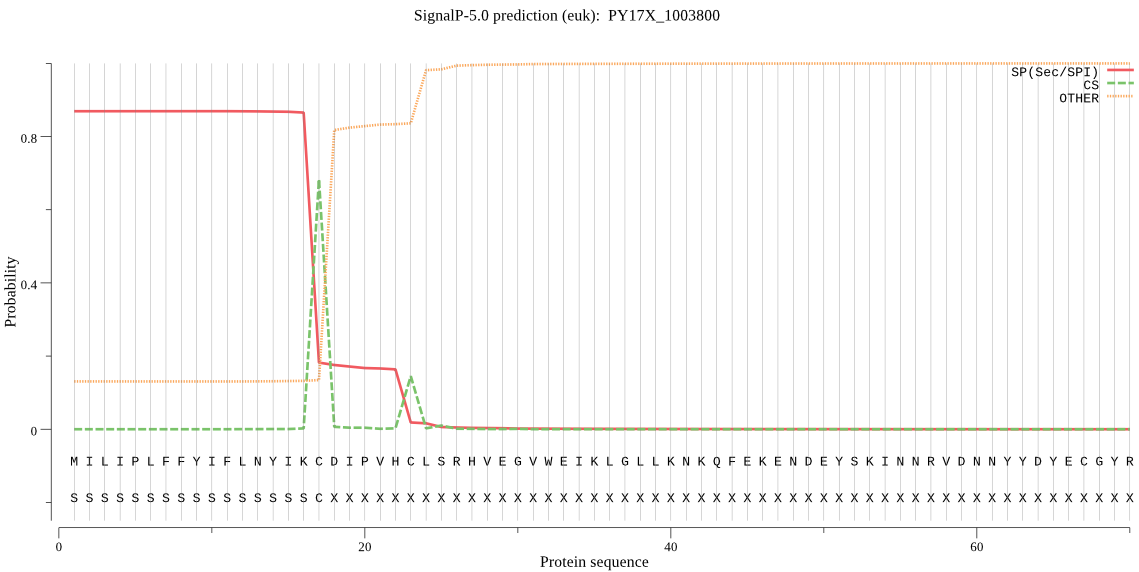
| PY17X_1003800 | SP | 0.056914 | 0.942817 | 0.000269 | CS pos: 17-18. IKC-DI. Pr: 0.9426 |
MILIPLFFYIFLNYIKCDIPVHCLSRHVEGVWEIKLGLLKNKQFEKENDEYSKINNRVDN NYYDYECGYRRPDDSNYHDALDPEIVKQRFEIKDKKIIAFNKDRTINIIENGKIVPEYSG YWRIVYDEGLHIEIYNSKNKNKEIYFSFFKFVKNGEVSYSYCNNLVMGVVSMYQLNDNTD IVKSNVNIVGNSRINNCNDNQSNSMVDDINVNSSNCVNKFEEKGKENNSSINNNITEKES GGKNGIFRYVYALYGVLKNSYLNILNNTVKVKENKSVALNDVDKLYGNNKDVKILIDHYN DNFIDFKTMPMKRYCWSGKKLDKNADKATNKISNMLSPLDVNADIQKHNYYINLIGDEKN KNILKNNKGKKRQTNNLQGVIDNPPTKKIKENSLFNMYSNRDITLKNFDWSDENDVKERL NGNIVRIFDEAIDQKECGSCYANSASRMINSRIRIKYSYIKKIDSLSFSNEQLLICDFFN QGCNGGYIYLSLKYAYENYLYTNKCFESYENLYINKDVKNNSLCDRFDTFKIFLKKKEKT NIYINNYPEKMEQKNEHVRMYNNFGNTHDNSNNRNDSNIHKKLIKGEDINYKYQSVDSGG QGKGNVSDDYLNEDYILVDKNMYEREKFKLDELDSCDTKIKVTKYEYLDIENEEDLKKYI YYNGPVAAAIEPSKSFIKYKKGILAGSFLKMQDGDKSNAYIWNKVDHAVVIVGWGEDTIE NLIKKKIKINNSKKKTHSYDEYESKNDDNLDEDIYDEHINSYVKNTKEKDKVVKYWKILN SWGTNWGYNGYFYILRDENYFNIRSYLLICDVNLFVKNKNVKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1003800 | 738 S | KKTHSYDEY | 0.997 | unsp | PY17X_1003800 | 738 S | KKTHSYDEY | 0.997 | unsp | PY17X_1003800 | 738 S | KKTHSYDEY | 0.997 | unsp | PY17X_1003800 | 204 S | NQSNSMVDD | 0.991 | unsp | PY17X_1003800 | 732 S | KINNSKKKT | 0.991 | unsp |