

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
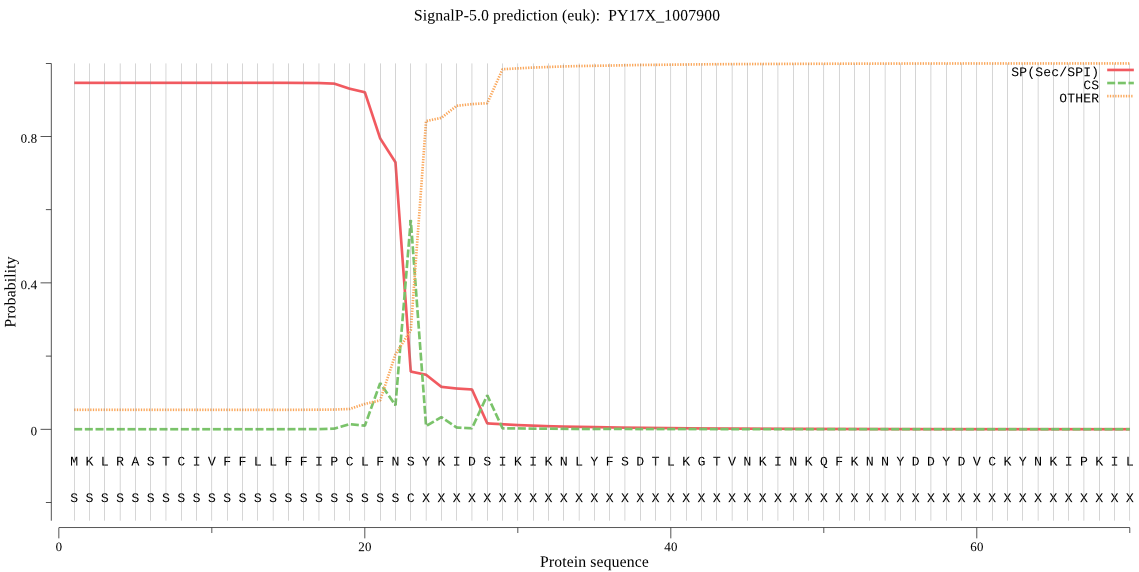
| PY17X_1007900 | SP | 0.032293 | 0.967559 | 0.000148 | CS pos: 23-24. FNS-YK. Pr: 0.4841 |
MKLRASTCIVFFLLFFIPCLFNSYKIDSIKIKNLYFSDTLKGTVNKINKQFKNNYDDYDV CKYNKIPKILIDRDKIKKNNKESGYIVGIENTCDDTCVCIIDLQLRIIKNFIISHFKVVH KYGGVYPFFISSINSVFLKHYVNKILEGIDASKIKCFGFSVCPGISQNMDTARNYIGDFK KRHKHIKASSINHVYAHVLSPLFFSFYNDKNTFTNNYEYKNESTQKDDHTDHINMNLFDT IKKDKIQMDKINSLLKVLNNNDVTKTKLKMITADDFLNYGSYVLKKKKIDDNNIQLDETK NNINQIKENTNMEKIPLNYIQNGYICVLVSGGSTQVYRVQKDKQNDINVCKISQTVDISV GDIIDKVARLLNLPVGLGGGPFLERESEKYIKTLNDQKINEDISFDLFEPFPVPFAPNNK INFSFSGIFNHLSKIIKELKKEKNFENEKSKYAYYCQKYIFKHLLNQLNKIMYCSELHFN IKNLFIVGGVGCNKFLFESLKKLALNRNKIENQLKEYIKLKKRLKKKIKKIENNNFLTKK NPPTNNMINEIELSSSFAWNFYLKNLLKKKTSNDILLALKAFDFNSFTKLKEKGSFLLQD QFLTSSITQPWNVYRTPINLSRDNAAMICFNTLLNMHNKINIHEDISEIKIKSTIKTQLQ NNFLLLSDIIVFDVFLDYFDHNVA
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_1007900 | 387 S | LERESEKYI | 0.996 | unsp |