

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
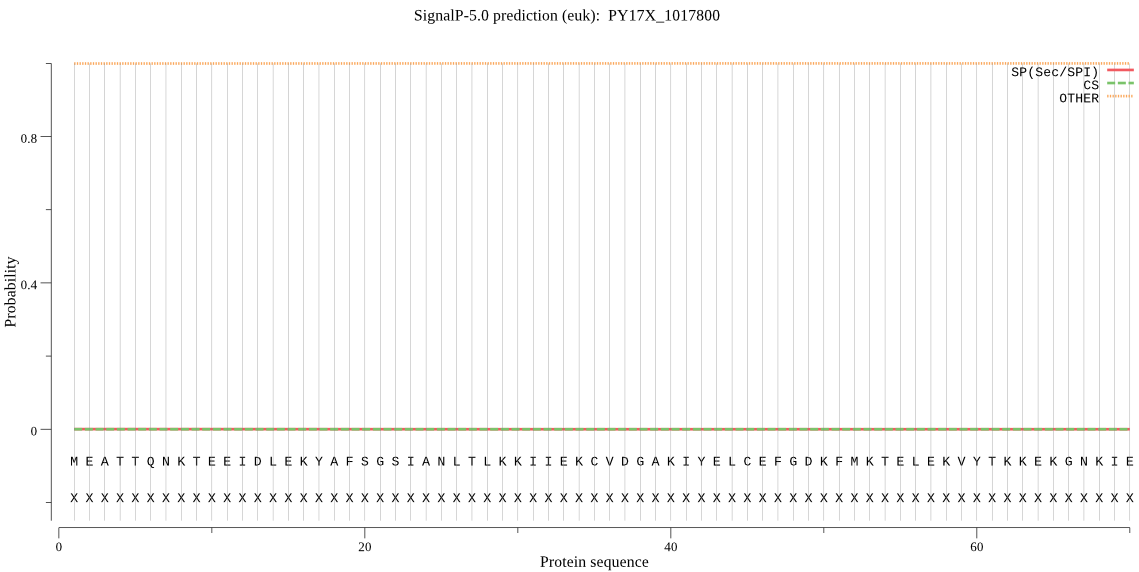
| PY17X_1017800 | OTHER | 0.999430 | 0.000539 | 0.000031 |
MEATTQNKTEEIDLEKYAFSGSIANLTLKKIIEKCVDGAKIYELCEFGDKFMKTELEKVY TKKEKGNKIEKGISFPVTINVNEVCNNYSPNADCDETIKNGDIVKISLGCHIDGHISIVG HTIYVGNENESIEGPKAEILKNSHILAQLFLKSLKVGAYASDITKNIQKACNDLKCNVIS NCVTYQIKKYILEGSKFILLKENPENKVEDFQIQPDDIYIIDVMVTTGDGKIKESDYKTT IYKREVQRNYQLKTNLGRSFINEINKNYPIFPFHSKFLEDQRASLIGIPEAMRHDLIKPY TVYTEKKKEYVSQFKYTVMVKEDGVKQFTGIKCPQINKCKTANTIQDEALKNILNSSGAK KKKDKLKNGEEATENAN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_1017800 | 131 S | NENESIEGP | 0.993 | unsp |