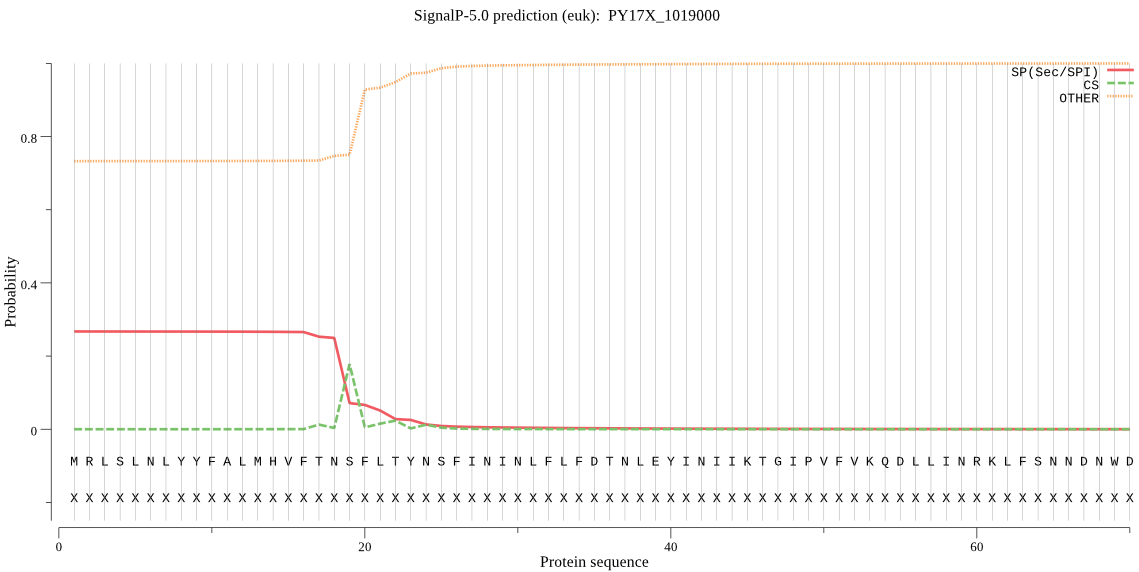
Fasta :-
MRLSLNLYYFALMHVFTNSFLTYNSFININLFLFDTNLEYINIIKTGIPVFVKQDLLINR
KLFSNNDNWDINDNTDNSVNEISTTRINVRKETEIYNINCKNINDEKCGIHAEGNQCMKK
NNNCITTLKLKTDKTPNEEAQISDNKIIDTDNTQIKKYVDSDKNIIKEKNEEKIMRKNDE
NVEMCKINNNENSGVELDTNKNMFDVRSNENCENYENCENCENVSTQKYTQIDDEILKIM
DKKNILVIINEEDENKSFHEEDDKEDDEEDDEEDDDELINKHLNEIENIINELSSKNYEQ
NKSTNSGDSDITDNTSNNLDDINEDEYEFVEGVCELDDINNVYVRYKNDLENYRNKLKEN
KLDEYYSDIIKSRNLININYENEYMLKMKYHKDYNFPLHNLHIVNEKYIKNNLYNNFVLL
NDENNNVSFQEINYYANLSKIDIRPTMFKEFYLKFSLCNGKYLPLGSSFVERVCFLSYHL
KQNPHLNKIKITKSKFILYHLFLNVIKAFKILSINKCVKHIKESNLLYIIFILNEEIIKE
SENEVNIGKCLNNIIERNKEWYEILNFIFTIMFGCSSYLKEHYSLTEEEKNNIFFKDNIL
DLPIFEYFNKNVIDIFIPRNIELKINIDLFKDISNSSDKNLKLYQTWYFCYLFIHKVYLL
NKMWNIIKKWETSNFVPNPWYIDHIGSVLVPHIISLRHIQQNDQKFFRYIDELQMFVSFK
RSKKYSIPNYDKNEFHLVEHIVAFQGTSSPFIWLFNLIYELTPYPFLTKGKVHKAYLYIF
KKAVKPYLHILKKKILNEIKNTKSKYSKENPYVIIFTGHSFGASMANMSSLYLENILREQ
GVKTKVNNNIDDIGNDDNDHNSVYIKIYSITFGMPIFYDDKYYEDFEKSAVISNNINVNY
DPIPVTMAIPILNGFRDPEENKKFNIIFKVEDLKKLNYDFGNIIFDGDELFNNEKNQPRS
ITFLFMKYFFKDNIFFELCEQHIYQTHYFFYFVFLTLLSRWANEAKWGTYFLIPFFIFDI
LSFSHKNVMFMLKENYKKYKKLVLKKTQSIEKQEKKQVAN