

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
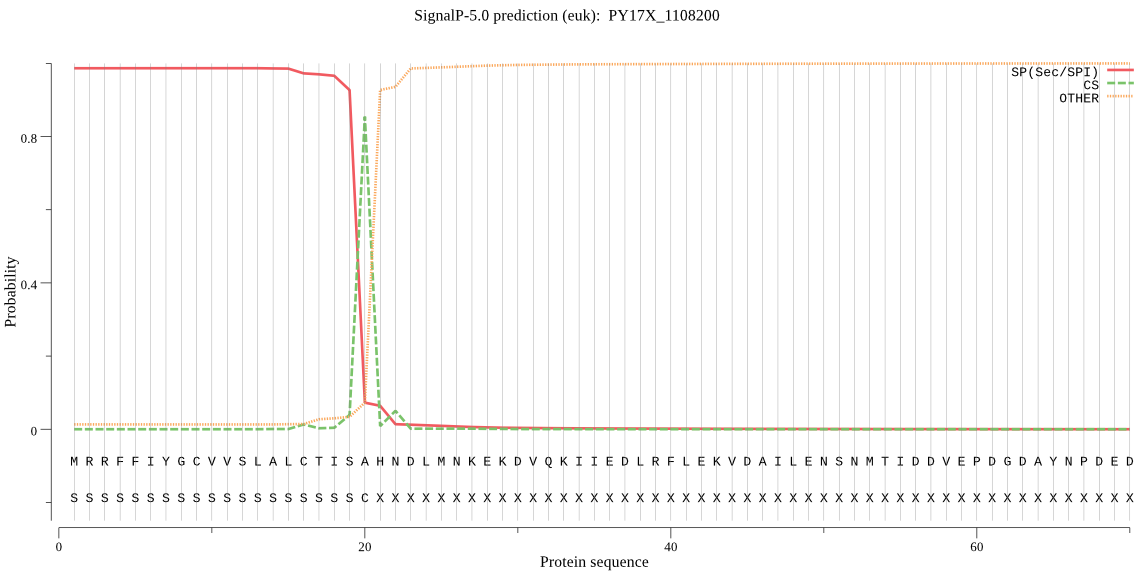
| PY17X_1108200 | SP | 0.006474 | 0.993128 | 0.000398 | CS pos: 20-21. ISA-HN. Pr: 0.7217 |
MRRFFIYGCVVSLALCTISAHNDLMNKEKDVQKIIEDLRFLEKVDAILENSNMTIDDVEP DGDAYNPDEDAPKEELNKIEMEKKKAEEEEKHSKKKILEKDLLNEKKNKSLRLIVSENHA TTPSFFEESIIQEDFMSFIQSKGEIVNLKNIKSMIIELNSDMTDKELETYITLLKKKGAH VESDELVGADSIYVDIIKDAVKRGDTSINFKKIQSNMLEVENNTYEKINNKLEKSKNSDK KSYFNDEYRNLQWGLDLARLDDAQEMITTNSVETTKVCVIDSGIDYNHPDLKGNIYVNLK ELNGKPGVDDDNNGIIDDIYGANYVNNTGDPWDDHNHGTHVAGIISAIGNNSIGVVGVNT NSKLVICKALDDKKLGRLGNIFKCIDYCINNKANIINGSFSFDEYSTVFSSTIEYLGRLG ILFVVSSSNCSHPTSSIPDITRCDLSVNSKYPSVLSTQYDNVVVVANLKKKKNGEYDVSI NSFYSDIYCQVSAPGANIYSTATRGSYLELSGTSMAAPHVAGIASIILSINPELTYKQVV SILKNSVVKLSSHKNKIAWGGYIDILKAVKNAISSKNSYIRFQGISIWKNKKRN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1108200 | 506 S | ATRGSYLEL | 0.993 | unsp | PY17X_1108200 | 506 S | ATRGSYLEL | 0.993 | unsp | PY17X_1108200 | 506 S | ATRGSYLEL | 0.993 | unsp | PY17X_1108200 | 93 S | EEKHSKKKI | 0.994 | unsp | PY17X_1108200 | 238 S | KSKNSDKKS | 0.995 | unsp |