

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
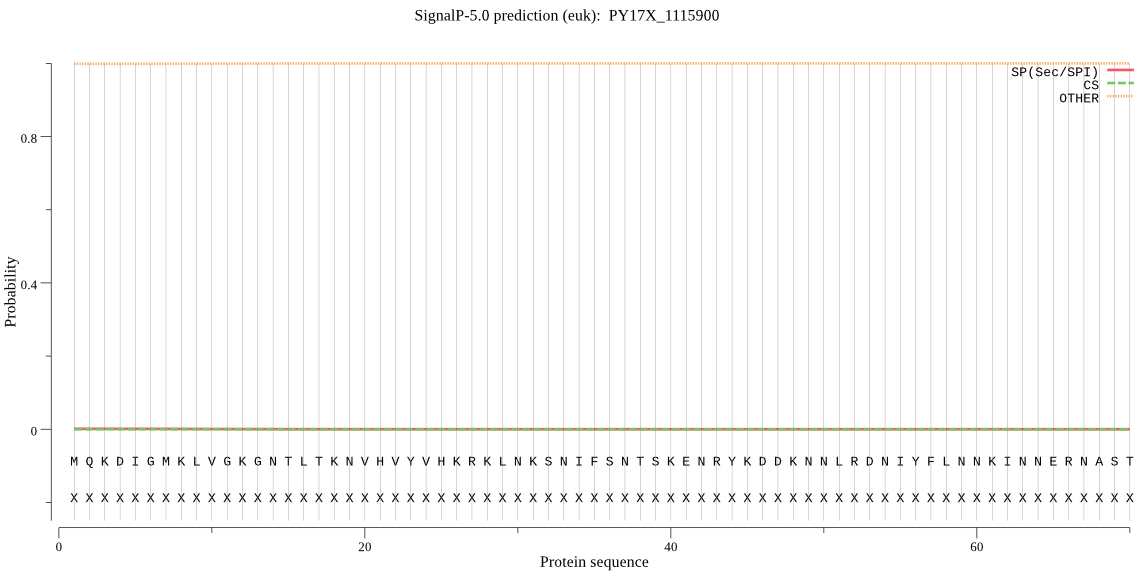
| PY17X_1115900 | OTHER | 0.988591 | 0.000023 | 0.011386 |
MQKDIGMKLVGKGNTLTKNVHVYVHKRKLNKSNIFSNTSKENRYKDDKNNLRDNIYFLNN KINNERNASTIYITANPSTSGFGKYVGNYSVRRFSKYSDKNIFNEQKRKCESLLFNFQNF EIKNYLLKLNKGRLQNSNIQKKIKNIIIKFEENYKNILYDKYSFFKKLCKEKSDFYKYRI SKINRNNYKTNLNFPINFLNIFISNNNNNNNKALKELINVGKYRGKLFLNNIPTYYKYGI KNIPYFKAVLMQHYNKSPVTYSLIFLHFFVYFLWINAKPDNMSYSYFSPAPIKSHSFSLL TSEFMYKYFCCSLKSLREKQLYTLVTNIISHNTIQSFLLNTISLFYIGRSLEILINSKNF FFTYIVSGIISSYIQILYQKNSSYGYKNVYVLGASGSISSILATYTFIHPNHKIYLYGVL GLPLALFSSLYFLNELYSIIANKNDNIGHASHLTGMFLGILYYYSYVNKKFRI
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1115900 | 95 S | VRRFSKYSD | 0.998 | unsp | PY17X_1115900 | 98 S | FSKYSDKNI | 0.991 | unsp |