

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
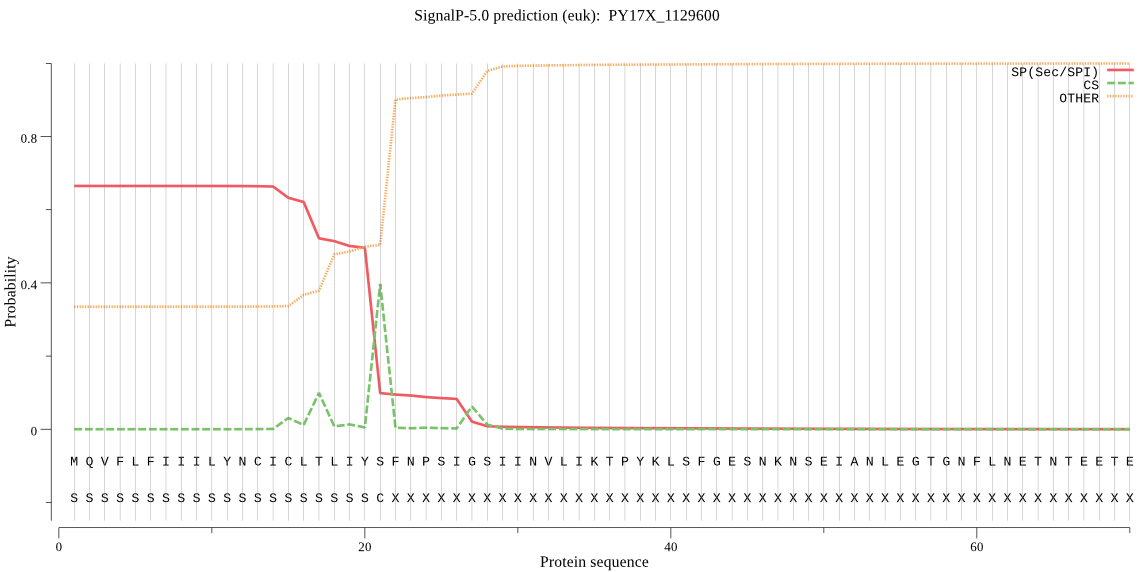
| PY17X_1129600 | SP | 0.049165 | 0.950499 | 0.000337 | CS pos: 21-22. IYS-FN. Pr: 0.8012 |
MQVFLFIIILYNCICLTLIYSFNPSIGSIINVLIKTPYKLSFGESNKNSEIANLEGTGNF LNETNTEETEQTVEFESKESKNDDQNVLNIESTPEAVVEYEVADTKEAIVEEASIDKETV IEETADKDDPSVEIKRVKEVTAEEIEKVKESAEKEIKKIKETAAEEVEKIKETVAEEIEI AKEIIAGAVLEQKQKEKEYVSEHTEENEEKKIESEYTEENEEKEIESKYTEENEEKKNES EYTEENEEDTVKEIEVSKNVNIENETIMEEIDNSDVEKLSETEKELKTNEMIQEKYGEFK RAEDSYYWESKSTDVEDGVDDDVDDDEIVGLPKKSNNTIIDNTPKSSVYLVPGLGGSTLI AEYNHAQIDSCSSKALHSKPYRIWLSLSRLFSIRSNVYCLFDTLKLDYDRKKKMYRNKPG VFINVEHYGYIKGVAFLDYIKNKPLRLTRYYGILADKFLENGYIDGKDILSAPYDWRFPL SQQKYEVLKSHIEYIYGLKKGTKVDLIGHSLGGLFINYFLSQFVDEEWKKKYINIVMHIN VPFAGSIKAIRALLYSSKDYTLFKLRNILKVSISGSLMKAISHNMGSPFDLIPYRKYYDR DQIVVIINMGKLPIDEKLVQSIVTECGIYNERCYTDREDVNLKTYTLSNWHELLSDDIRE KYENYIQYTDRFFSVDHGIPTYCLYSTTRKRNTEYMLFYQDVHFNHDPIIYYGIGDGTIS LESLEACKKLQNVKEAKHFVYYKHIGILKSDIVSDYIYNIINQNRTN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1129600 | 556 S | ALLYSSKDY | 0.993 | unsp | PY17X_1129600 | 556 S | ALLYSSKDY | 0.993 | unsp | PY17X_1129600 | 556 S | ALLYSSKDY | 0.993 | unsp | PY17X_1129600 | 80 S | ESKESKNDD | 0.992 | unsp | PY17X_1129600 | 280 S | VEKLSETEK | 0.994 | unsp |