

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1131900 | OTHER | 0.999938 | 0.000026 | 0.000035 |
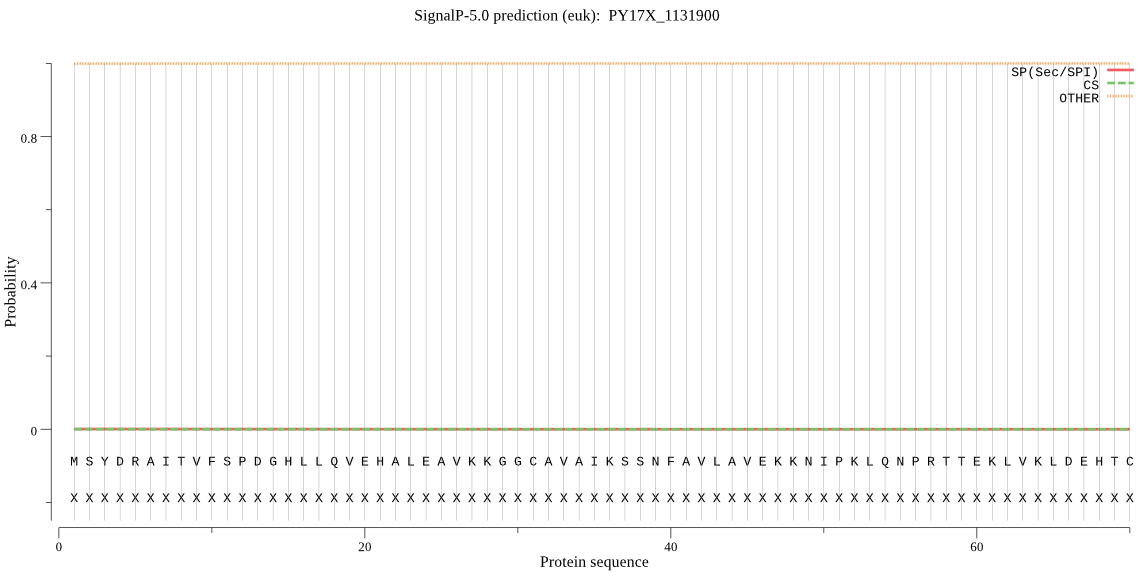
MSYDRAITVFSPDGHLLQVEHALEAVKKGGCAVAIKSSNFAVLAVEKKNIPKLQNPRTTE KLVKLDEHTCLAFSGLNADARILVNKTRLECQRYYLNMDQPAPVDYIAKYVARIQQKFTH RGGVRPFGIATLITGFKNNKEICIYQTEPSGIYASWKAQAIGKNAKVVQEFLEKNYQENM EKNECLLLALKAIFEVVELSSKNVELALLTEEELKYIDEQEVNSLVEIIDNERTKKNAQN E
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_1131900 | 59 T | NPRTTEKLV | 0.992 | unsp |