

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1136100 | SP | 0.025813 | 0.972827 | 0.001360 | CS pos: 16-17. ARG-NS. Pr: 0.5405 |
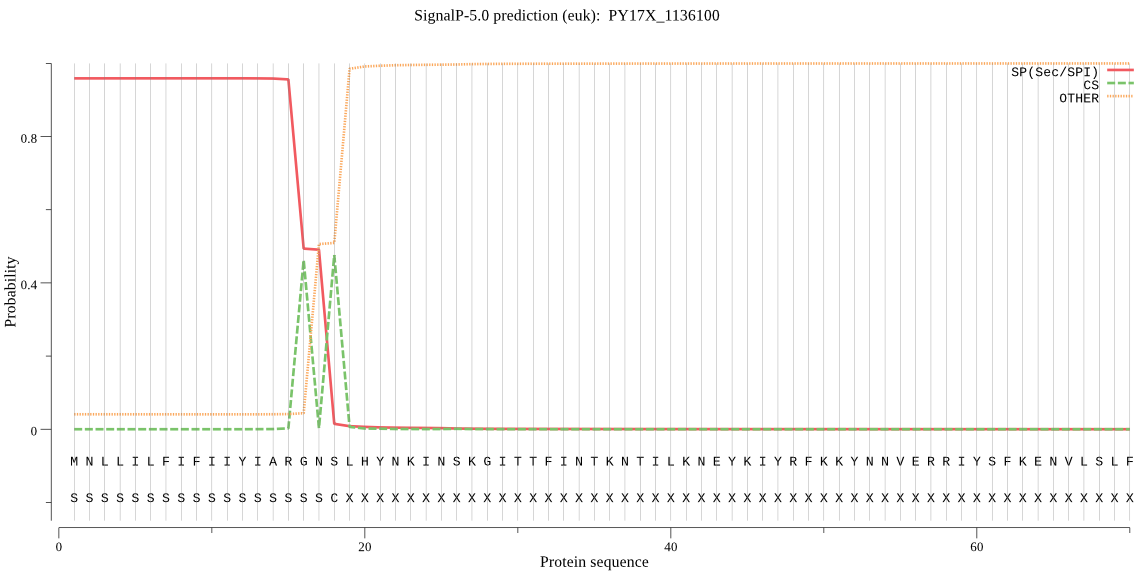
MNLLILFIFIIYIARGNSLHYNKINSKGITTFINTKNTILKNEYKIYRFKKYNNVERRIY SFKENVLSLFNTVKDKEKITNLLENISNNVKNKFPNRFTYYNYLFNKCKLDRILIVINTL LYLYLNRVDKNEEKKIFFTKGNLVQIKDEQKAEKYQCNYYDIYKNKNYKTLFSSIFIHKN ILHLYFNMSSLMSIYKMVSPIYSNSQILITYLLSGFLSNLISYIYYIKPQKKNIFLKDII DQNYYSRNTPLNKPNKIICGSSSAIYSLYGMYITHMIFFYIKNNYIVNTGFLYNIFYSFL SSLLLENVSHFNHVLGFMCGFVMSSTLILFDNN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PY17X_1136100 | 61 S | RRIYSFKEN | 0.996 | unsp |