

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1225700 | SP | 0.075574 | 0.924357 | 0.000069 | CS pos: 25-26. ITC-NK. Pr: 0.8080 |
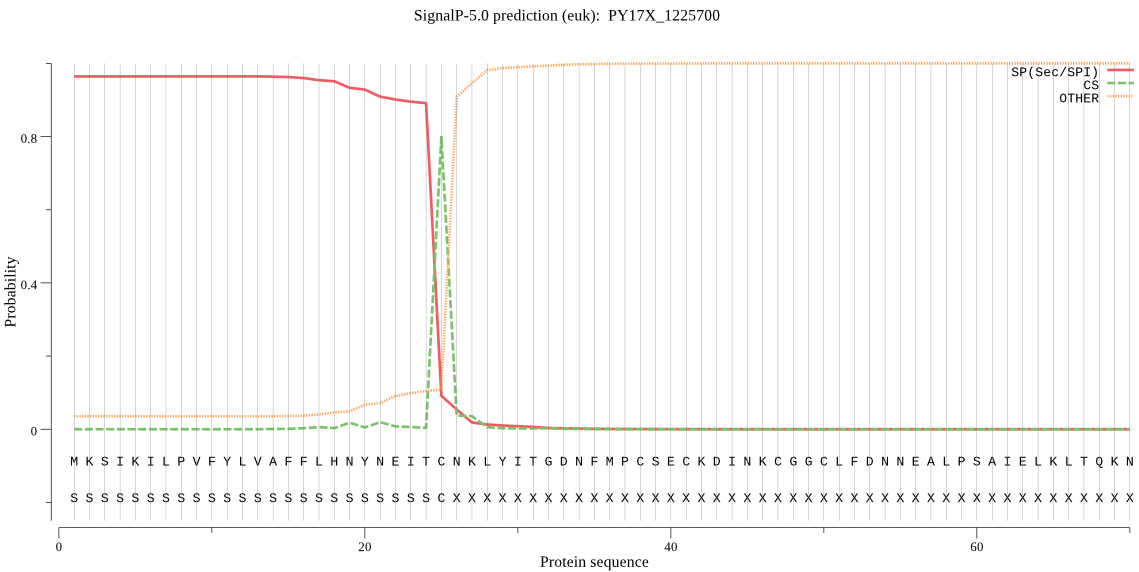
MKSIKILPVFYLVAFFLHNYNEITCNKLYITGDNFMPCSECKDINKCGGCLFDNNEALPS AIELKLTQKNRDNNNHDNLKIHHDSLKLGNVKYYVKRGEGVSGSFGNVSGNDINSMAEIH NEIKNRKEKKEENKSSLSFIDHSNNNNSEEGKGDYNFSKIQKHEQDGDKINTQEEFEKIQ SQAVNKSGVSFSDRVLDENGNQAQPSKGVTIEETSDNVFLVPLQHLRDSQFVGKLLVGTP PQEIHPIFDTGSTNLWVVTTECKEDSCKKVHQYNPNKSKTFRRSFIKQNLHIVFGSGAIT GTLGKDNFILGNHIIRNQTFGLVKSETSDNLNNSDNVFEYINFEGIVGLGFPGMLTAGNI PFFDNLLKQYKNMTPQFSFYISPNDETSTFIVGGISKSYYEGDIYMLPVIKEYYWEVKLD AIYIGDEKICCEEESYAIFDSGTSYNTMPSTQINNFFKIVSSKPCNEENYNDILKEYPSI KYVFGKLVIELLPNEYMIVNDDLCVPAYMQIDVPSENNNAYLLGTIAFMRHYFTIFVRGQ EGNPSMVGVAKAKRV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1225700 | 398 S | GISKSYYEG | 0.994 | unsp | PY17X_1225700 | 398 S | GISKSYYEG | 0.994 | unsp | PY17X_1225700 | 398 S | GISKSYYEG | 0.994 | unsp | PY17X_1225700 | 436 Y | EEESYAIFD | 0.99 | unsp | PY17X_1225700 | 138 S | KSSLSFIDH | 0.994 | unsp | PY17X_1225700 | 382 S | SFYISPNDE | 0.992 | unsp |