

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
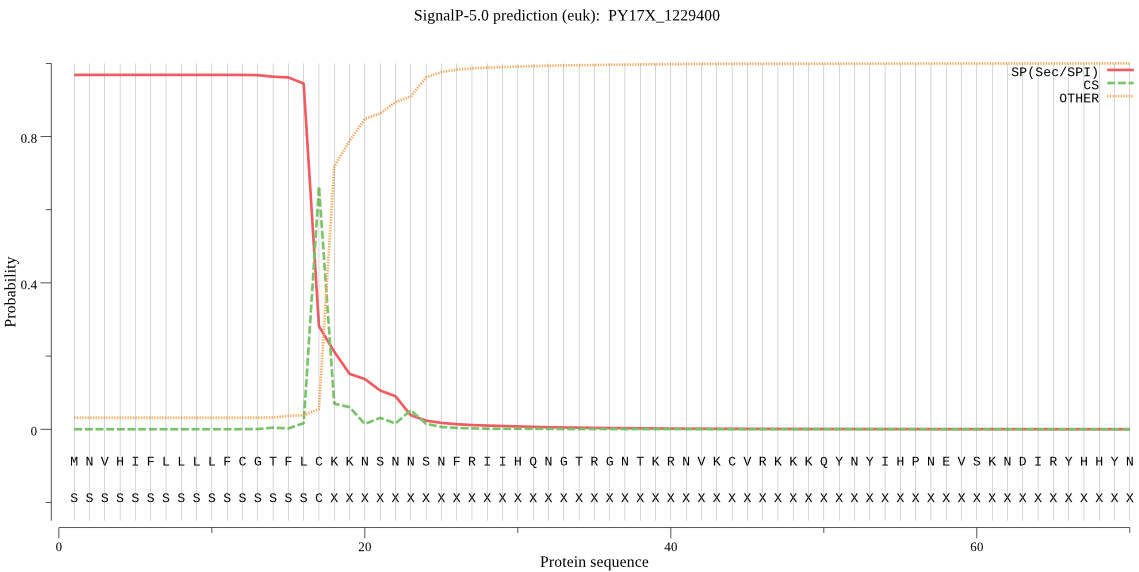
| PY17X_1229400 | SP | 0.016490 | 0.981824 | 0.001686 | CS pos: 17-18. FLC-KK. Pr: 0.7491 |
MNVHIFLLLLFCGTFLCKKNSNNSNFRIIHQNGTRGNTKRNVKCVRKKKQYNYIHPNEVS KNDIRYHHYNFFITTHTHKKKLAKKDKFKRRYNLNSSGEGFSNTIHNNVKKNTNNYSEEQ LPSYHKYIENFKTRKIIHPSIRITDHDKKFMKCKESYNKLYSHLTETELFENFSYVGRQR KGILSPKYYLPKYIDRPNYHKTGTPIYVNYENNLKSKEHISIRNDDNNNNETDEKYHKTY FTTNKYEYNNVKSDYDIEIISRNCKFARELMDDISYIICEGITTNDIDIYILNKCVNNGF YPSPLNYHLFPKSSCISINEILCHGIPDNNVLYENDIVKVDISVYKDGYHADMCESFIVQ KISKEEKKKRKKNYDYIYLNDKLRTKHTKYIIKYNFDLTTHKIVKKGKYPSVRKVRYNPP NSKEHENEYIEEYDDNTNSVFGENSYNNNYYTYNDDPNNVSENYEDELENFHRRYDEDFI FNPKKGELYNNLQQYIYQKGMEKDKKGKEMNRKFEFFDTNKQDITNMKKFMFEKNRDLIK TAYDCTMAAISICKPGVPFKNIAKVMDDYLKKKNNSYQYYSIVPNLCGHNIGKNFHEEPF IIHTLNNDDRKMCENLVFTIEPIITERSCDFITWPDNWTLSNSRYYYSAQFEHTILITKN GAKILTQKTETSPKYIWEND
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1229400 | 253 S | NNVKSDYDI | 0.994 | unsp | PY17X_1229400 | 363 S | VQKISKEEK | 0.996 | unsp |