

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1230000 | OTHER | 0.985268 | 0.003043 | 0.011689 |
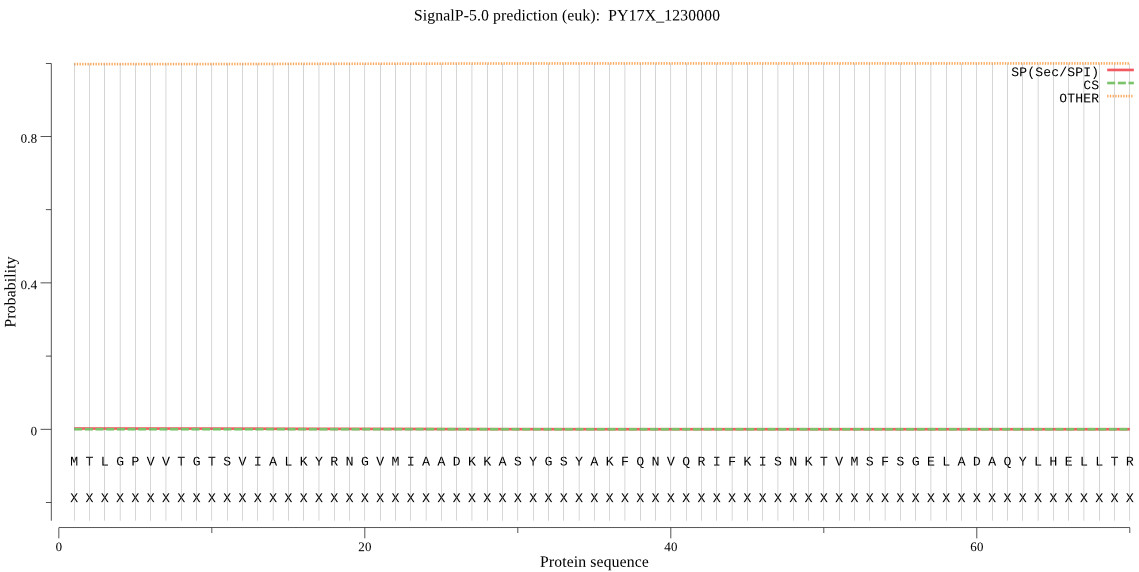
MTLGPVVTGTSVIALKYRNGVMIAADKKASYGSYAKFQNVQRIFKISNKTVMSFSGELAD AQYLHELLTRVNVNDVVEKKTKYDLHDTKFYHSYVSRLFYNRKNKIDPLFNNIIIAGLNS QEYDDNDKDILLYSEKQNNEEYKDIDKKDLYIGFVDMHGTQFCEDYITTGYARYFALTLL RNHYKDNMTEDEARSLLNECLKILYFRDTTASNKIQIVKVTSKGVEYEEPYILNCDLNSR DYIYPSTMLPATGCMW