

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1319800 | SP | 0.048212 | 0.951622 | 0.000166 | CS pos: 18-19. IIG-IK. Pr: 0.4813 |
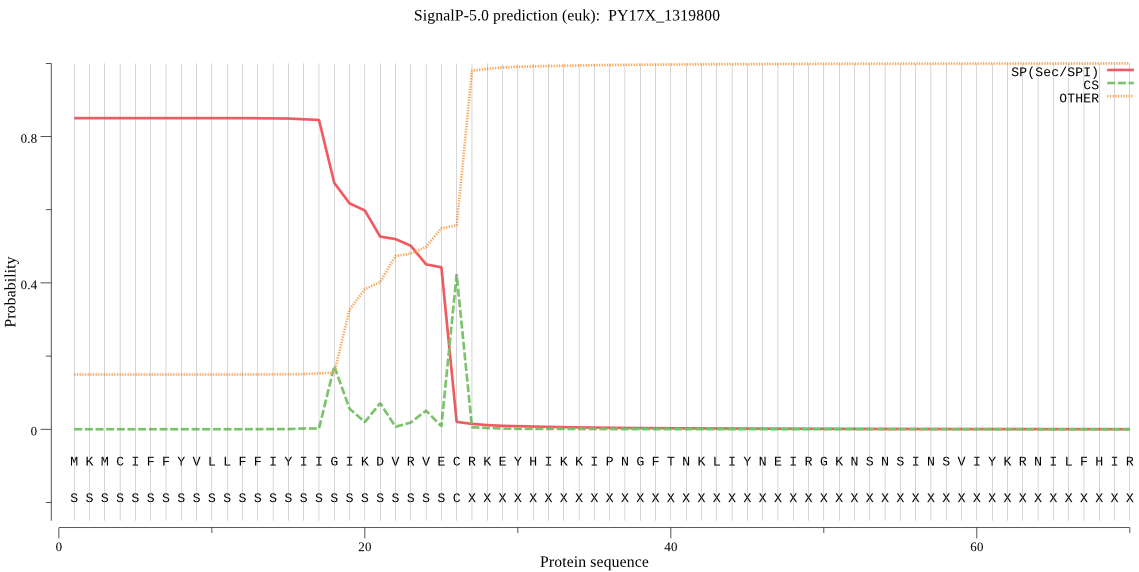
MKMCIFFYVLLFFIYIIGIKDVRVECRKEYHIKKIPNGFTNKLIYNEIRGKNSNSINSVI YKRNILFHIRIPKKTLQGRNLFSNDIRNKKINEFLQKKKNSCYLHINSNVNEIKKKRDKI LKNVFLNAKLLQEYFNNFKSSLLYKYKNTKIITKLFLSTSFLVMILNVFGLRPEDIALHD KRIIRAFEFYRIITSALFYGDISLYVLTNIYMLYLQSQELEKSVGSSETLAFYLSQITIL SAICSYIKKPFYSTALLKSLLFTNCMLNPYNKSNLIFGINIYNMYLPYLSIVIDILHAQD FKASLSGILGIISGSIYYLSNIYLLEKCNKKFFKIPQILRNYLDSFNTDEFVF