

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
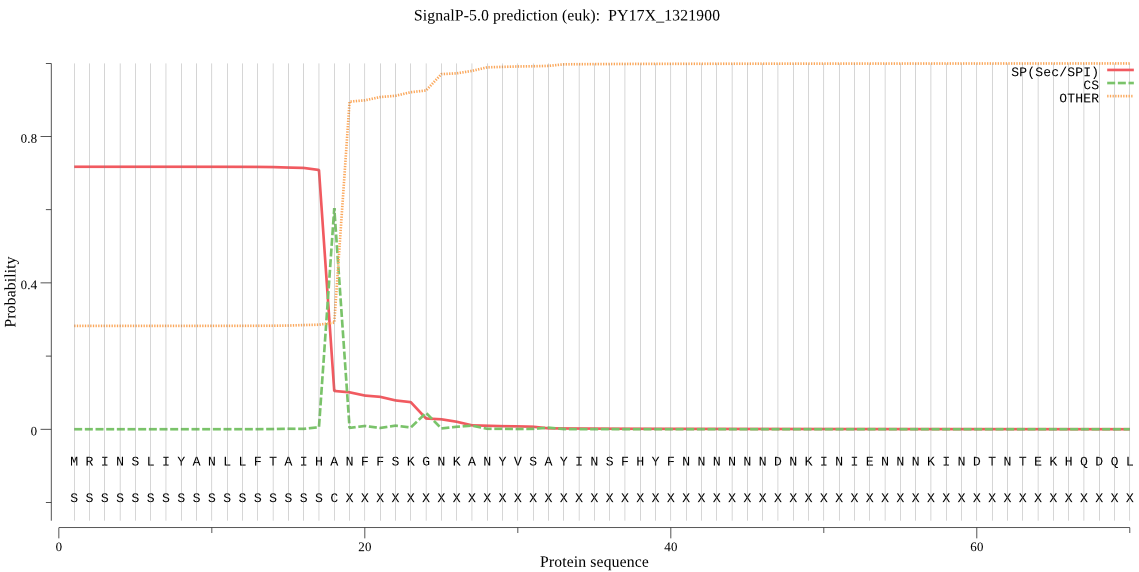
| PY17X_1321900 | SP | 0.052596 | 0.921861 | 0.025543 | CS pos: 18-19. IHA-NF. Pr: 0.9601 |
MRINSLIYANLLFTAIHANFFSKGNKANYVSAYINSFHYFNNNNNNDNKINIENNNKIND TNTEKHQDQLNNFFTRRFYSLYSKNRYLREGKKNSKEVNLNNDNTNNYNMESSNEQNNQM MGGDSYKERLQNLKKYMGDHNIDVYIIINSDAHNSEIINDQDKKIYYLTNYSGADGILIL TKDAQIVYVNSLYELQANKELDTNFFTLKIGRITNRDEIFQTIVDLKFNTIAFDGKNTSV SFYEKLKNKIKIQFPDKKIQEKFIYKNSINQVVKDNNINLYVLETPLVTVPNNDVNKKPI FIYDREFGGSCAAQKIQESSDFFIENPDVDSLLLSELDEIAYLLNLRGYDYKYSPLFYSY VYLKYNRDKGRIDDIILFTKVENVQKNVLAHLERIHVKLMDYDSVVSYLTKNVSSKSENT KNNNGKNIILGSIHENSSPRYDISLSPHINLMIYMLFNKEKVLLKKSPIVDMKAIKNYVE MDSIKEAHVLDGLALLQFFHWCEEKRKTKELFKETEISLRNKIDYFRSTKKNFISLSFST ISAIGPNSAIIHYESTEDTNAKITPSIYLLDSGGQYLHGTTDVTRTTHFGEPTADEKKLY TLVLKGHLSLRKVIFASYTNSMALDFLARQALFNNFLDYNHGTGHGVGICLNVHEGGYSI SPAAGTPLKENMVLSNEPGYYWADHFGIRIENMQYVVTKKQTDNAKFLTFNDLTLYPYEK KLLDYSLLTPEEIADINEYHQTIRNTLLPRIKENPSDYAKGIEQYLMDITEPIHTKQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1321900 | 483 S | VEMDSIKEA | 0.995 | unsp | PY17X_1321900 | 483 S | VEMDSIKEA | 0.995 | unsp | PY17X_1321900 | 483 S | VEMDSIKEA | 0.995 | unsp | PY17X_1321900 | 555 S | IHYESTEDT | 0.997 | unsp | PY17X_1321900 | 125 S | MGGDSYKER | 0.995 | unsp | PY17X_1321900 | 415 S | KNVSSKSEN | 0.995 | unsp |