

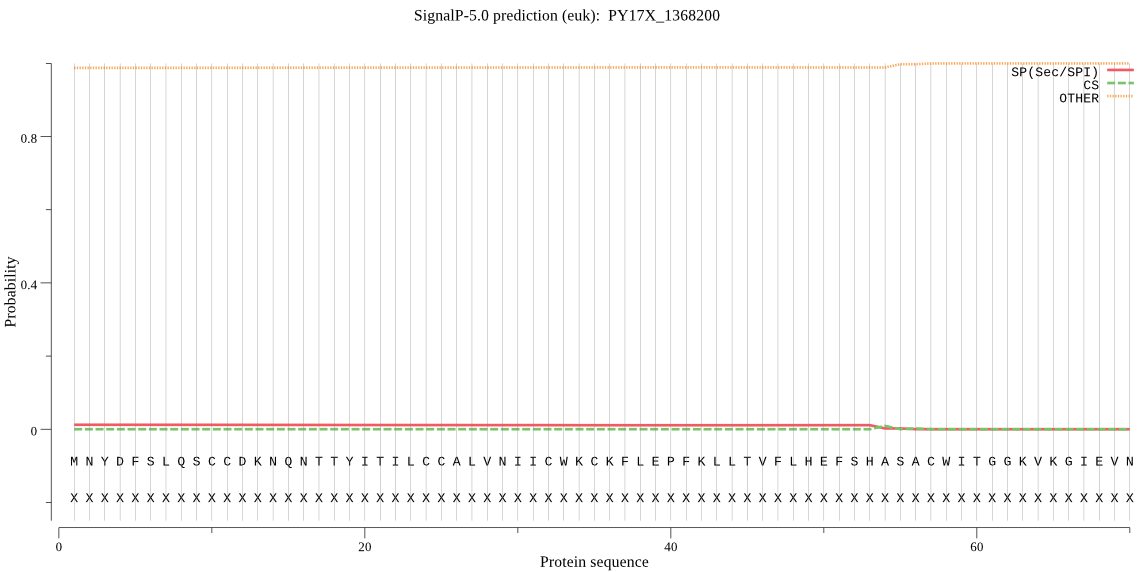
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1368200 | OTHER | 0.999981 | 0.000014 | 0.000006 |
MNYDFSLQSCCDKNQNTTYITILCCALVNIICWKCKFLEPFKLLTVFLHEFSHASACWIT GGKVKGIEVNRDHGGCTNTIGGDMFFILPAGYIGSCFYGMFFILMAYINKWTLLASAVFL CFLLLIVLTFYAKNFFLRFLCILFLGIIIGTWGIGVSFNESKYWPLKVIMTFMGVLNEMY SMVDIFDDLITRTTPESDAYKYAKLTKCNSKFCGVLWCVINLGFIILTMYLIGAIHVKPF EN