

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
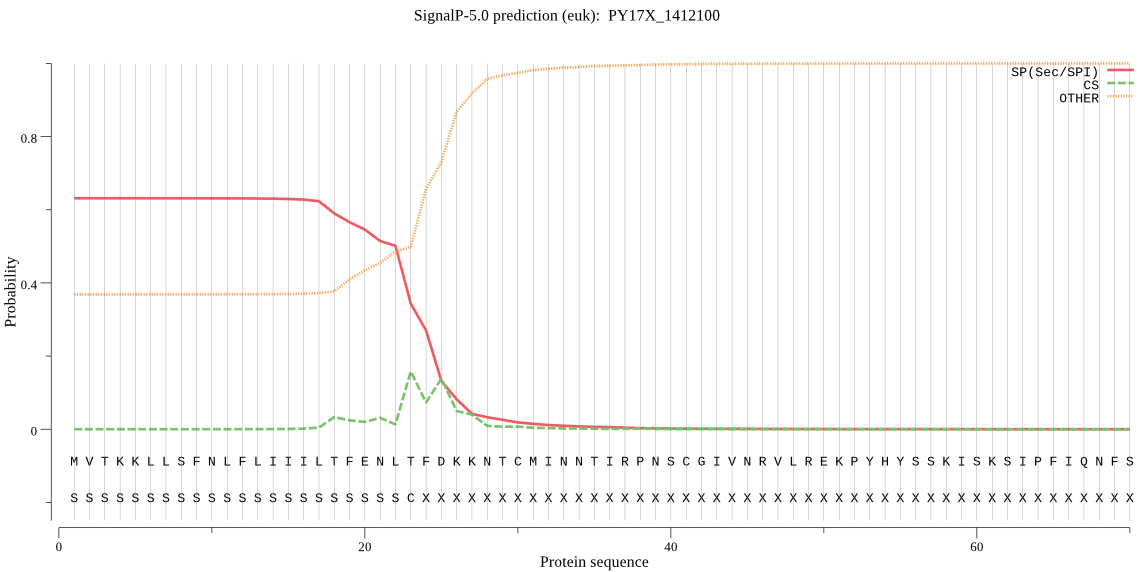
| PY17X_1412100 | SP | 0.133388 | 0.865660 | 0.000952 | CS pos: 23-24. NLT-FD. Pr: 0.2491 |
MVTKKLLSFNLFLIIILTFENLTFDKKNTCMINNTIRPNSCGIVNRVLREKPYHYSSKIS KSIPFIQNFSLEKNFTGESLQKNILNNINKLGGAHLFHISKSHLAAKAGNKNTEFIGEAT ELFKGFKRNFGINMTENKQTNIGRMLCEDDNNNGGEVTSTEKTIFKNSKDPQIHYRTDYK PSGFTIDNVTLNINIFDNETIVRSSLNMCTNENYADEDLVFDGVGLSIKEISINNNKLNE GEDYTYDNEFLTIFAKNVPKENFVFLSEVVIHPETNYALTGLYKSKDIIVSQCEATGFRR ITFFIDRPDMMAKYDVTLTADKTKYPVLLSNGDKLNEFDIPGGRHGARFNDPHLKPCYLF AVVAGDLKHLSDNYVTKYTKKPVELYVYSEAKYVSKLKWALECLKKAMKFDEDYFGLEYD LSRLNLVAVSDFNVGAMENKGLNIFNADSLLASKKTSIDFSFERILTVVGHEYFHNYTGN RVTLRDWFQLTLKEGLTVHRENLFSEETTKTATFRLTHIDLLRSVQFLEDSSPLSHPIRP ESYISMENFYTNTVYDKGSEVMRMYQTILGDEYYKKGIDIYLKKHDGGTATCEDFNDAMN EAYQMKNGNTDENLDQYLLWFSQSGTPHVTAEYIYDENEKTFTINLSQITYPDDNQKEKY PLFIPVKVGFISPKDGKDVIPEVVLELKKDKESFVFQNVSEKPIPSLFREFSAPVYIKDN LTDEERIALLKYDSDAFVRYNVCIDLYMKQIIKNYNELVSQKTKENDLVELSLTPVNDEF INAIKHLLEDKHADPGFKAYIIALPRDRYIMNYIKEVDPIVLADTKDYIYKQIGSRLNPI LFSIFQNTESKANDMTHFKDESYIDFDQLNMRKLRNSILVMLSKAQYPHMLKYIKEQSKS AYPSNWLASLSASAYFSGDDYYDLYDKTYKLSKNDELLLQEWLKTVSRSDRSDIYSIIKK LEVEILKDSKNPNNIRAVYLPFTANLRAFNDISGKGYKLMADVIMKVDKFNPMVATQLCD PFKLWNKLDLKRQALMHDEMNRMLSMENISPNLKEYLLRLTNKM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1412100 | 672 S | VGFISPKDG | 0.998 | unsp | PY17X_1412100 | 672 S | VGFISPKDG | 0.998 | unsp | PY17X_1412100 | 672 S | VGFISPKDG | 0.998 | unsp | PY17X_1412100 | 1045 S | NRMLSMENI | 0.995 | unsp | PY17X_1412100 | 227 S | GVGLSIKEI | 0.991 | unsp | PY17X_1412100 | 542 S | IRPESYISM | 0.992 | unsp |