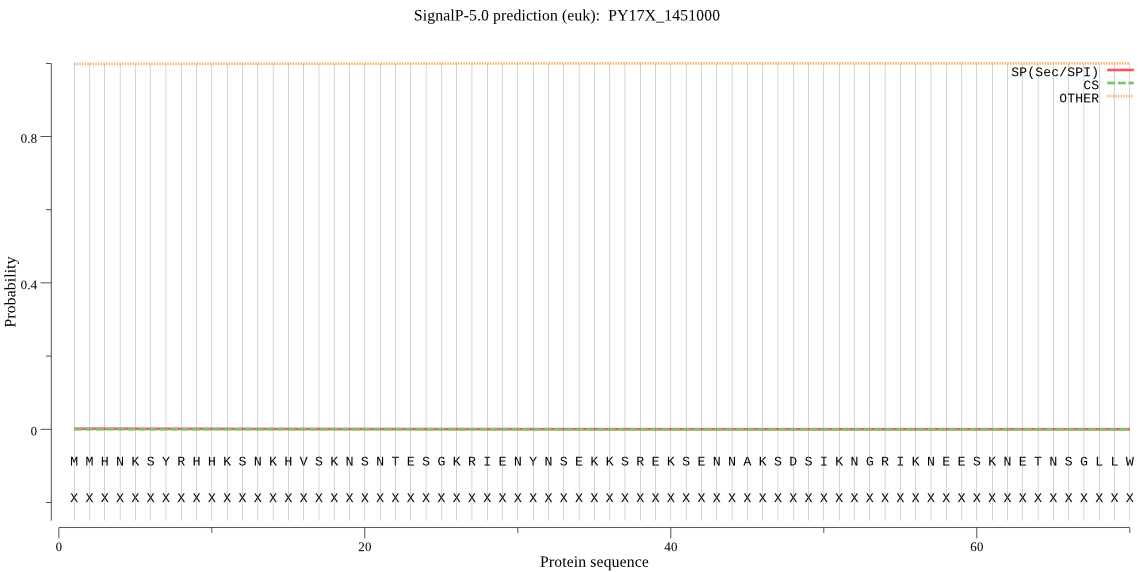
Fasta :-
MMHNKSYRHHKSNKHVSKNSNTESGKRIENYNSEKKSREKSENNAKSDSIKNGRIKNEES
KNETNSGLLWSCVKNLYSSFSSAVKNKIFINDIIEENNRKHNKNIVSDKFRLIDKKSKKY
KEKGLNRNKYSISVISNDRCSSIDAHHKNKSFISNKIKENYKQKYEKHEKHEKHEKHEKY
EKHEKYEKHEKYDKHDKHDKHDRHDKYKRDEENVSQKKKKKKKDFTYEFSENDIKNLFMS
SNDILEYPYYQSIQNDLTKNDENNNLPFRNIINDLVHKDLDVAHKELDAVHKELDAVHKE
LDVTHKELDVVAKYSDSKSTITNNNIFKENNLSSYVTKNASENSQLHNFDLKNKNILNEL
IKNKQNEIKIKEERQDDISINNKFENKNINNLYNCTINLKTNYSHLSFETLHNYEHNVKI
SEIENETNISFNKNNEIENETNISFNKNNEIENETNISFNKNNEIENETNISFNKISYSQ
KTNLISSIQHSDINVMHQLLEDHGITNVGEKDAKYKEFDKKSGEKIGEKIDKKIGERIGE
KIGEKIGENEQAKATMFSERENEKKNKEMHNVGDQKSNAANNNNNNQPSFLDSLFYSKQN
EDEDVIILSDIKPNNSLSDEKISYQKEIHAKRVIGNDNNDGDNDDNNCGNNGDNDDNNCG
NNGDNDYAWSEIPLPNKINKPNNKLREQNVNDYIKNNEYMKKIRNQYKSLIRVIDTYIDD
NINGEGLNNSFINKNEKNSLNDIEENIIDSSIDRCNNESIKIIEDIKLGNEDKYVLLKYD
EDSLIEALEKLRIDKQKEIEKKNKKEKIDKNIFFKFKQKNYYDEAICILNKKSDNNILIE
KFNVPLMYSQIKCLIDTCWLNDEIINFYLSMLQEYNETSIKNGLTNFPKMFTFSTFFFQS
LNFNGSYNYNKVSRWTKRKKINILEYDLILIPLHVGGNHWTLGAISIKNKHIKLYDSLNM
PNKKFFEYMRRYIVDEVKDKQQITIDISPWTYDSNGLPESGIPCQENGYDCGVFTCMFAK
CLTFNRDFDFDQKDIKEIRLKMVYEISQGHLVF