-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs: GO:0003860
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
|
_ID | Prediction | OTHER | SP | mTP | CS_Position |
|
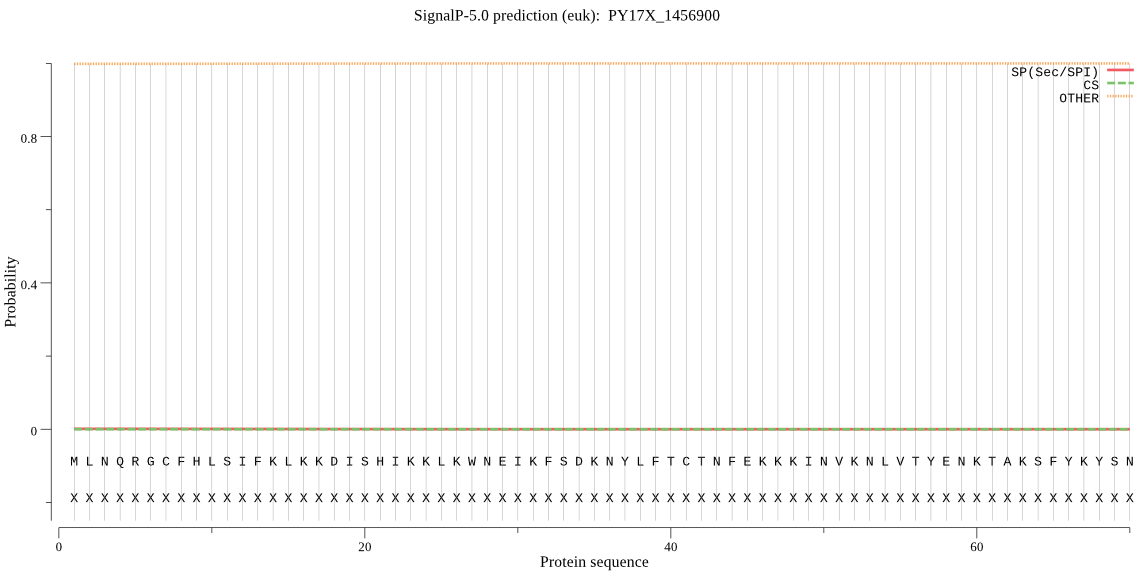
PY17X_1456900 | OTHER | 0.853056 | 0.000063 | 0.146881 | |
No Results
-
Fasta :-
>PY17X_1456900
MLNQRGCFHLSIFKLKKDISHIKKLKWNEIKFSDKNYLFTCTNFEKKKINVKNLVTYENK
TAKSFYKYSNNSNLHTNAHKIENNHKNVLSKKMENTNDNINMNKRFISDECNKGDLFKKT
EKEKNNELDKYNMENMDDVTKSKNGNCNDIIDLTLSDVWSKKTLIVEYKNNIVEILLNRK
EKLNAINKDMINGLLNMVKSLSNDDRCNLIVIRSINQNCFCSGSDVKDIVQNKDQGINHL
KQLYKYINFISKINKNILCIWNGYAMGGGLGISMYTKYRIINKNVIFAMPENKIGFFPDV
GSSYFLKKYFPRNIALHLGLTSLRLNEIDLINFKVCTNYIENLDQFLNELYNIKKEDPNK
FNEELIKILNKYPPKVNVNTKPVLTEELISNIDKYYTSANNLEELINNLKKNEDNDNFCK
QLLLDINANCYFSCQLWFSYFIYNYDKPMEEVLDNDFKMTQYFLYHTNTFEKGVTEILIK
KNKSFQWSKDHENNPVKLEETIEDILMNKNLLSIKDEFI
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PY17X_1456900.fa
Sequence name : PY17X_1456900
Sequence length : 519
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.988
CoefTot : -0.504
ChDiff : 11
ZoneTo : 17
KR : 4
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.424 1.106 0.224 0.615
MesoH : -0.422 0.284 -0.374 0.248
MuHd_075 : 21.532 15.402 8.054 4.023
MuHd_095 : 12.321 5.091 5.054 1.438
MuHd_100 : 9.886 7.860 2.945 1.934
MuHd_105 : 8.385 8.959 1.966 2.828
Hmax_075 : 1.700 9.000 -0.730 2.910
Hmax_095 : 3.300 5.800 -0.094 2.180
Hmax_100 : 0.300 5.200 -1.244 2.410
Hmax_105 : 0.500 9.100 -2.158 2.660
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.7748 0.2252
DFMC : 0.6917 0.3083
-
Fasta :-
>PY17X_1456900
ATGCTAAATCAAAGAGGATGTTTTCATCTCAGTATATTCAAATTAAAAAAAGATATTTCA
CATATAAAAAAATTAAAATGGAATGAAATAAAATTTAGTGATAAAAACTATTTGTTTACA
TGTACAAACTTTGAAAAGAAAAAAATAAACGTGAAAAATTTGGTGACTTATGAAAATAAA
ACCGCAAAAAGTTTCTATAAATATTCTAATAATAGTAATTTACATACTAATGCACATAAA
ATAGAAAATAATCATAAAAACGTATTATCTAAAAAAATGGAAAATACAAATGACAATATT
AATATGAACAAGCGATTTATATCTGATGAATGTAATAAAGGCGACCTTTTTAAGAAAACT
GAAAAAGAGAAAAATAATGAATTAGATAAATATAATATGGAAAATATGGATGATGTTACA
AAATCAAAGAATGGTAACTGTAATGATATTATAGATCTAACCCTATCTGATGTATGGTCA
AAAAAAACATTAATAGTAGAATATAAAAACAACATAGTCGAAATATTATTAAATAGGAAA
GAAAAATTAAATGCTATAAATAAAGATATGATTAATGGATTATTAAATATGGTAAAAAGT
TTAAGCAATGATGATAGATGTAATTTGATAGTAATAAGAAGTATCAATCAAAATTGTTTT
TGTTCGGGTTCAGATGTAAAAGATATAGTTCAGAATAAAGATCAAGGGATAAATCATTTA
AAACAACTATATAAGTATATTAATTTTATTTCAAAAATTAATAAAAATATCTTATGCATA
TGGAATGGGTATGCTATGGGTGGGGGATTAGGTATATCTATGTATACAAAGTATCGAATC
ATAAATAAAAATGTAATATTTGCCATGCCAGAAAATAAAATAGGTTTTTTCCCTGATGTT
GGAAGCTCTTATTTTTTAAAAAAATATTTTCCACGAAATATTGCTTTACACTTAGGATTA
ACATCATTGCGATTAAATGAAATTGATTTAATTAATTTTAAAGTATGTACTAATTATATA
GAAAACCTAGACCAATTTTTGAATGAATTGTATAATATTAAAAAAGAAGACCCAAATAAA
TTTAATGAGGAACTTATTAAAATTTTAAATAAATACCCCCCAAAAGTTAATGTTAATACA
AAACCAGTATTAACAGAAGAATTAATTAGTAATATCGACAAGTATTATACATCTGCCAAT
AATTTAGAGGAATTGATAAATAATTTAAAAAAAAATGAAGATAATGATAATTTTTGTAAA
CAATTATTATTAGATATAAACGCTAATTGTTATTTTAGCTGCCAATTATGGTTCTCTTAT
TTTATATATAACTATGACAAACCTATGGAAGAAGTATTAGATAACGATTTTAAGATGACT
CAGTATTTTTTATATCACACTAACACATTTGAAAAGGGGGTAACTGAAATATTAATAAAA
AAAAATAAAAGCTTTCAATGGAGTAAGGACCATGAAAATAACCCTGTGAAATTGGAAGAA
ACTATTGAAGATATTTTGATGAATAAAAATTTATTGTCTATTAAAGATGAATTTATATAA
- Download Fasta
- title: substrate binding site
- coordinates: L183,A185,N218,S222,G223,S224,D225,V226,Y264,M266,G267,G268,P290,E291,I294
|
ID | Site | Peptide | Score | Method |
|
PY17X_1456900 | 513 S | KNLLSIKDE | 0.994 | unsp |


