

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PY17X_1463300 | SP | 0.000465 | 0.999531 | 0.000004 | CS pos: 19-20. AKG-DL. Pr: 0.9220 |
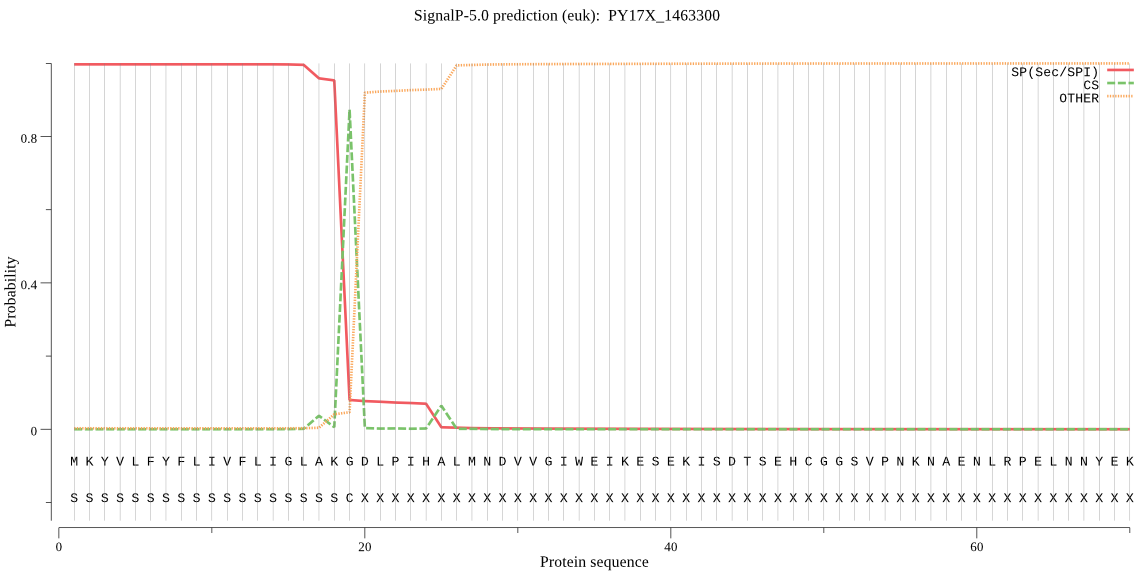
MKYVLFYFLIVFLIGLAKGDLPIHALMNDVVGIWEIKESEKISDTSEHCGGSVPNKNAEN LRPELNNYEKYLQTNYGELKNLKINLTFEKIKLFNDSSSRGIWTYLAVRSVDDNNSIIGN WTMVYDEGFEIRLRGKRYFGFFKYDKKVSEECPSIVENINKINTECYKTDPTKIRLGWIF YERKQKNDKEKIYQWGCFYAEKITKMPISSFVIHNMNNSNKEDNNFVNNKEHTDTDTNFL GLMQKDNYNKIRMNHPGLFGSTKISYIDRHGGKERRIYGCKKRESKKMEMNLVLPKEFSW GDPYNNKNFDEIVEDQKECGSCYLISSVYVLEKRFEILLSKKYKKNIKMNKLFYECIINL SQYNQGCDGGFPFLVGKEIYENGIHATMNYGSLDNKISSKIEKFSRNKKNKEIYYASDYN YISGCYECSSEYDMMREILNNGPIVAAIYATSSLLKIYKLNDYNFIYTTISDENKICDVP NKEFNGWQQTNHAVVIVGWGEYINKNNKLIKYWLIRNTWGNHWGYKGYLKYQRGINLNGI ESQAVYIDPDFTRGSGKDLLV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PY17X_1463300 | 555 S | FTRGSGKDL | 0.997 | unsp | PY17X_1463300 | 555 S | FTRGSGKDL | 0.997 | unsp | PY17X_1463300 | 555 S | FTRGSGKDL | 0.997 | unsp | PY17X_1463300 | 149 S | DKKVSEECP | 0.992 | unsp | PY17X_1463300 | 285 S | KKRESKKME | 0.997 | unsp |