

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0113800 | SP | 0.205503 | 0.751920 | 0.042578 | CS pos: 22-23. IRG-GG. Pr: 0.4738 |
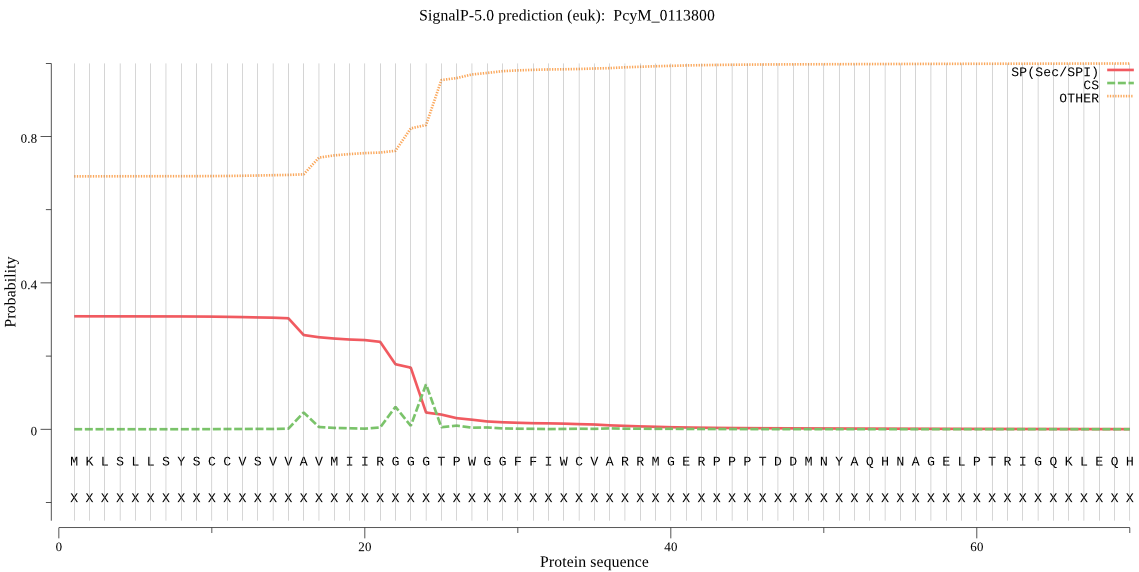
MKLSLLSYSCCVSVVAVMIIRGGGTPWGGFFIWCVARRMGERPPPTDDMNYAQHNAGELP TRIGQKLEQHAHMGTETKGDDSEFIFNKSTPTDGSKKINVDLPLVKYAPAYVNSLLNDKQ VAKMGAPSGENRLSRSDQHMGGDLHRGGDLHRGGDLHRGGDHHPNDITKGERFALSKQQH EASYRGNAEGGEYASERNNQGGEQPPYGTPNTWRRKKKKNLHYRMNPTEHHLMDYSLMKD KQIHRGETLNSYVNNIDDHNSKRRMQPRVGKLKLRINRDDKKTIRDTLTPQVVKVMKEYL LKGRKKYRNGQMDHLGGISSDQLDRRDGTGWGSSWSGSSKVNRVLRFFPANFAPHSGDNQ RGAAKRSIINEKLHRNLKQITLGAAGSGDADGERAEDAKKGAKDAKDAKDIKDARDAKNN AEAGALHNIYRGVVKLYVDITEPSLEMIWSNSPPKRVTGSGFVIEGDLILTNAHNVAYST RILVRKHGSSKKYEATVLHVAHEADIAILTVSDRSFYQDVSALELGPLPSLRDDVITVGY PSGGDKLSVTKGIVSRIEVQYYKHSNERLLLTQIDAPMNPGNSGGPALVKGKVAGICSQL LKTANNTSYIIPTPVIKHFLLDLHKSGKYNGYPSLGVKYVPLDNANLRRLVGLTDLEKGK AVEENSGILVTEVDEEQMRYQPADVDYCAYAEGATQAEVSTGQASSTGAHALATGVDPFP RRPPTDAHCYGLKKNDVIISIDGTQIKSDGTATLRGDESVDFQFLLNEKYVQDLCTCRVV RKGRIKSVVVRVSRVNYLVRQHNRDVRNKFFIYGGVIFTTLTRSLYPDEDTDNVELLRLL QFNLFKKRRGDEIVIVKRILPSKLTIGFNYTDCIVLTVNGIPVRSLQHLVEVIDKRDGPA GGEAANMANAANMANAANTANTASTANTKEVPSTSGNANPYLMSTLENDPFLHVQEKDNL LHFLLLTSNGQQVPLVLLRNEVQQESAEIKRVYGITRERYLYNG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0113800 | 542 S | VGYPSGGDK | 0.994 | unsp | PcyM_0113800 | 542 S | VGYPSGGDK | 0.994 | unsp | PcyM_0113800 | 542 S | VGYPSGGDK | 0.994 | unsp | PcyM_0113800 | 134 S | ENRLSRSDQ | 0.998 | unsp | PcyM_0113800 | 490 S | KHGSSKKYE | 0.992 | unsp |