

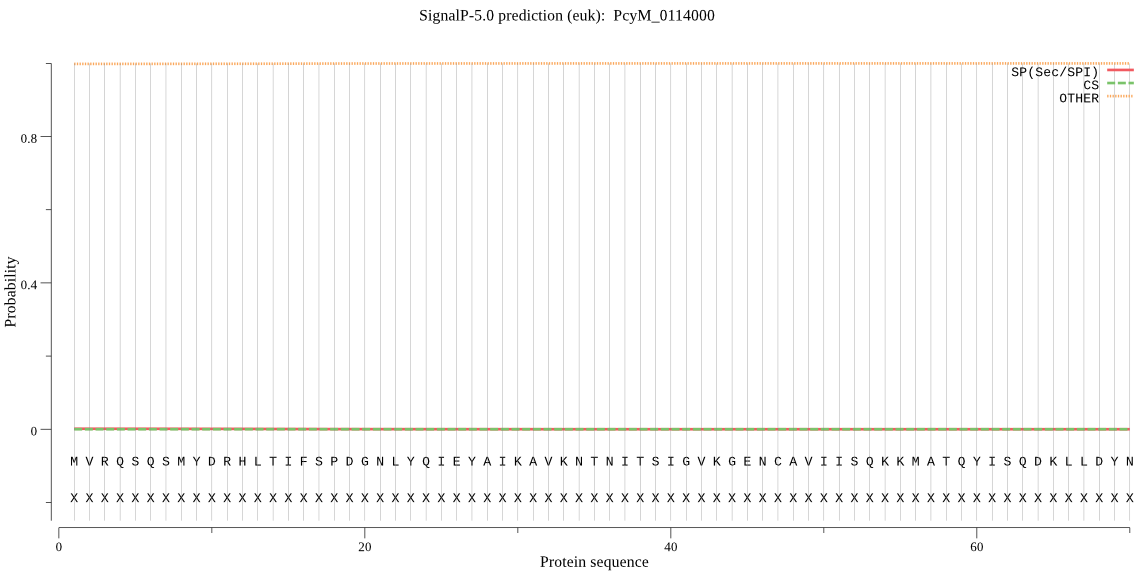
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0114000 | OTHER | 0.999419 | 0.000300 | 0.000281 |
MVRQSQSMYDRHLTIFSPDGNLYQIEYAIKAVKNTNITSIGVKGENCAVIISQKKMATQY ISQDKLLDYNNITNIYNITDEIGCSMVGMPGDCLSMVYKARIEAAEFLYSNGHNVNVETL CRNICDKIQVFTQHAYMRLHACSGMIIGMGEDNKPGLFKFDPSGFCAGYRACVIGNKEQE SISILERLLEKRKKKIQQETLEEDVQNTIILAIEALQTILAFDLKANEIEMAIVSKANPN FTQISEKEIDNYLTFIAERD