

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0117400 | OTHER | 0.999564 | 0.000407 | 0.000029 |
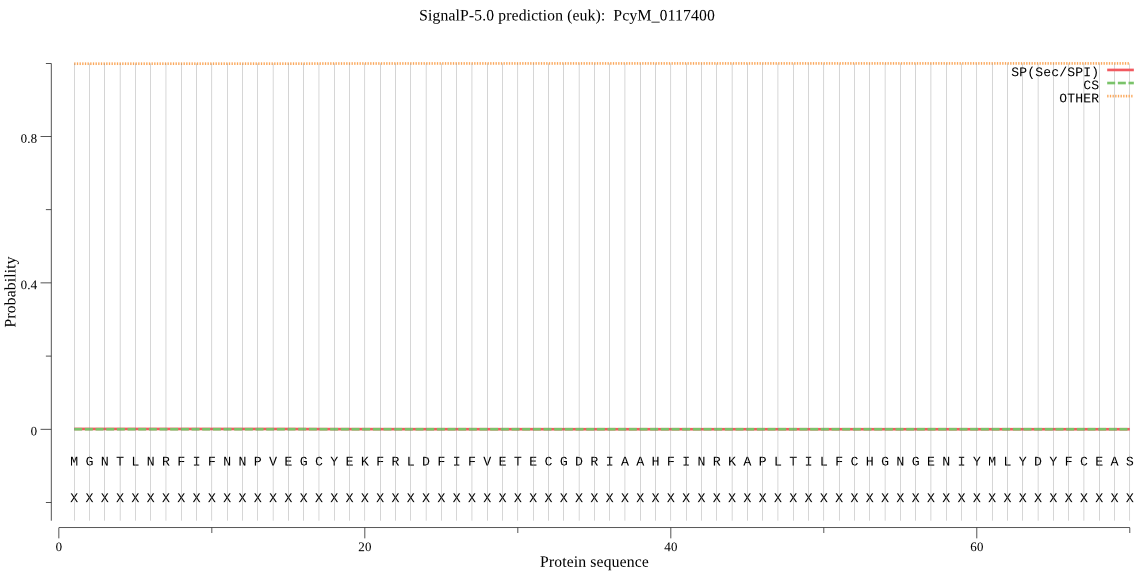
MGNTLNRFIFNNPVEGCYEKFRLDFIFVETECGDRIAAHFINRKAPLTILFCHGNGENIY MLYDYFCEASKIWNVNVFLYDYPGYGESTGTPNEMSMYQSGRAVYDYMVNVLNIKAESIV LYGKSIGSCAAIDIAIVRKVKGIILQSALMSLLNICFKTRFILPFDSFCNIKKIGMVPCF AFFIHGTDDKIVPFYHGLSLYEKCKLKVHPYWVVGGKHNDIELIENKKFNDSIKSFLKFL RNV