

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
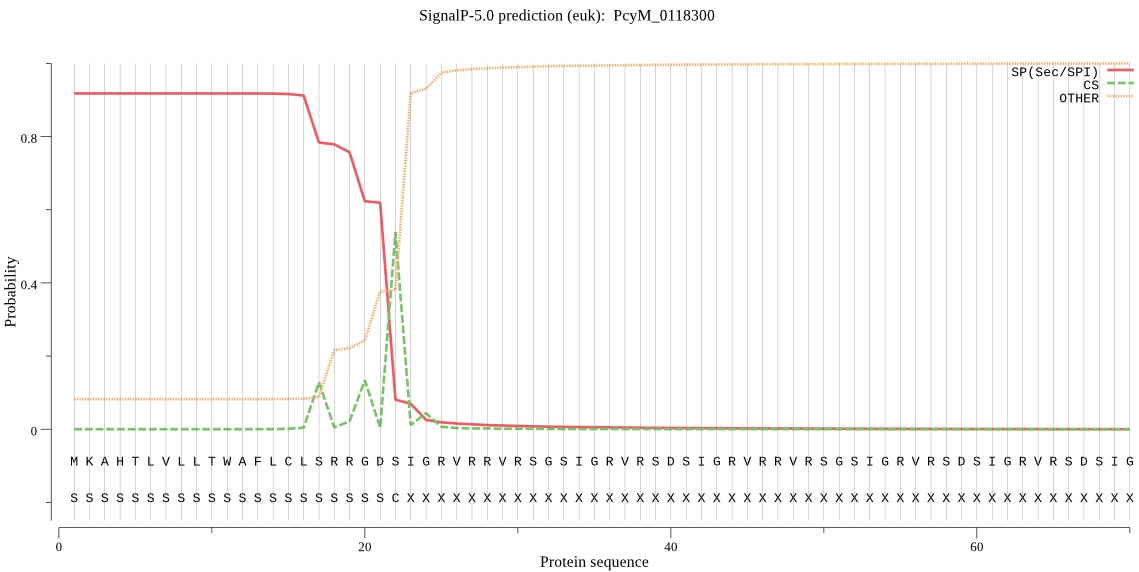
| PcyM_0118300 | SP | 0.014810 | 0.983707 | 0.001483 | CS pos: 22-23. GDS-IG. Pr: 0.7512 |
MKAHTLVLLTWAFLCLSRRGDSIGRVRRVRSGSIGRVRSDSIGRVRRVRSGSIGRVRSDS IGRVRSDSIGRVRSGSIGRVRSGSCADAARTRPRRGFIHLGKHSKGSYAKQRFPHHLNST TAGEGFSNTVHHDVKKNTQNYSENSPPASFHRYIENMKTRKNVHPSIRITEFDKKFIKCK ESYNSRYSHLKDSELFGSFHFVGRQKKGIMSPKYYLPKYVERPNYHRTGTPVYVPYEGEG QVSSSTKGRKSPYPYANIKSEEDIETIRSNCQFARELMDDVSHIICEGVTTNDLDIYILN KCINNGYYPSPLNYHLFPKSSCISINEILCHGIPDNNVLYENDIVKVDISVFKDGFHADM CESFLVPKLTKNEKKRKKKFYDFIYLNDKLRTRYTKFILKYNFDLITNKIVKTGKQCSIR RIKYDPPNSKDRPSEQYHSYDDNTNSVLRSGHFGGYAATHHEDYYSDAGGAAHLGGEDLE RDNFERDDLAGDHFERDDLDDLELFHRQYDEHVIYNKRQSSQTYDDIQKYIYRKTREPQN NQNNQNRFAFFDKRKLDVDELKIYMHKKNMDLIRTAHECTMEAIRVCKPGVPFSAIGDAI ADHLDRKNNQHVRYAVVPHLCGHNIGKNFHEEPFIIHTRNDDDRRMCENLVFTIEPIISE RACDFIMWPDNWTLSNARYVFSAQFEHTIVVRKDGAEILTGKTDRSPKFVWECEQE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0118300 | 41 S | VRSDSIGRV | 0.996 | unsp | PcyM_0118300 | 41 S | VRSDSIGRV | 0.996 | unsp | PcyM_0118300 | 41 S | VRSDSIGRV | 0.996 | unsp | PcyM_0118300 | 52 S | VRSGSIGRV | 0.992 | unsp | PcyM_0118300 | 60 S | VRSDSIGRV | 0.996 | unsp | PcyM_0118300 | 68 S | VRSDSIGRV | 0.996 | unsp | PcyM_0118300 | 76 S | VRSGSIGRV | 0.992 | unsp | PcyM_0118300 | 82 S | GRVRSGSCA | 0.992 | unsp | PcyM_0118300 | 84 S | VRSGSCADA | 0.996 | unsp | PcyM_0118300 | 245 S | QVSSSTKGR | 0.994 | unsp | PcyM_0118300 | 251 S | KGRKSPYPY | 0.998 | unsp | PcyM_0118300 | 260 S | ANIKSEEDI | 0.993 | unsp | PcyM_0118300 | 439 S | EQYHSYDDN | 0.996 | unsp | PcyM_0118300 | 22 S | RRGDSIGRV | 0.993 | unsp | PcyM_0118300 | 33 S | VRSGSIGRV | 0.992 | unsp |